Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
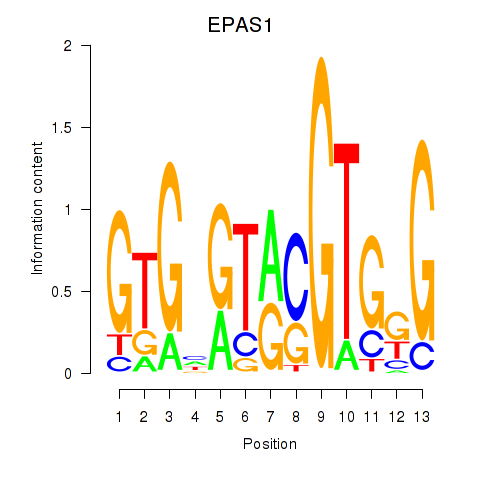
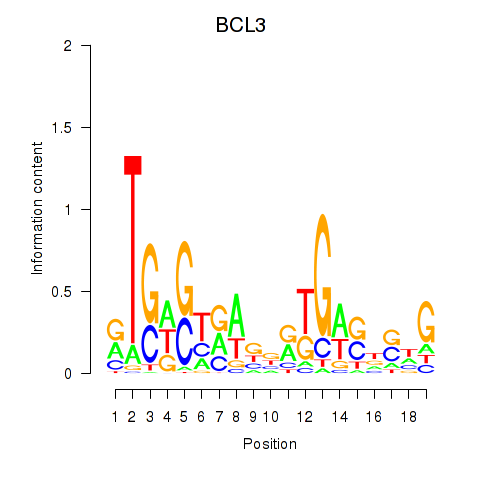
Results for EPAS1_BCL3
Z-value: 0.28
Transcription factors associated with EPAS1_BCL3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EPAS1
|
ENSG00000116016.9 | endothelial PAS domain protein 1 |
|
BCL3
|
ENSG00000069399.8 | BCL3 transcription coactivator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL3 | hg19_v2_chr19_+_45254529_45254578 | 0.52 | 4.8e-01 | Click! |
| EPAS1 | hg19_v2_chr2_+_46524537_46524553 | -0.11 | 8.9e-01 | Click! |
Activity profile of EPAS1_BCL3 motif
Sorted Z-values of EPAS1_BCL3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_238475217 | 0.29 |
ENST00000165524.1
|
PRLH
|
prolactin releasing hormone |
| chr20_+_34680698 | 0.14 |
ENST00000447825.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr19_+_45445491 | 0.13 |
ENST00000592954.1
ENST00000419266.2 ENST00000589057.1 |
APOC4
APOC4-APOC2
|
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr2_+_33701684 | 0.12 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr13_+_113030658 | 0.12 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr22_-_46283597 | 0.11 |
ENST00000451118.1
|
WI2-85898F10.1
|
WI2-85898F10.1 |
| chr5_+_52776228 | 0.10 |
ENST00000256759.3
|
FST
|
follistatin |
| chr9_+_2017063 | 0.10 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_-_7080227 | 0.09 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr17_-_77179487 | 0.09 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr15_+_74466012 | 0.09 |
ENST00000249842.3
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr11_-_414948 | 0.09 |
ENST00000530494.1
ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr9_+_34652164 | 0.09 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr17_-_41623009 | 0.08 |
ENST00000393664.2
|
ETV4
|
ets variant 4 |
| chr7_-_140340098 | 0.08 |
ENST00000477488.1
|
DENND2A
|
DENN/MADD domain containing 2A |
| chr3_-_47484661 | 0.08 |
ENST00000495603.2
|
SCAP
|
SREBF chaperone |
| chr22_+_42372970 | 0.08 |
ENST00000291236.11
|
SEPT3
|
septin 3 |
| chr4_-_105416039 | 0.08 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chrX_-_142722897 | 0.08 |
ENST00000338017.4
|
SLITRK4
|
SLIT and NTRK-like family, member 4 |
| chr5_+_52776449 | 0.08 |
ENST00000396947.3
|
FST
|
follistatin |
| chr7_-_29235063 | 0.08 |
ENST00000437527.1
ENST00000455544.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr16_+_57662419 | 0.07 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr22_-_24096562 | 0.07 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr17_-_191188 | 0.07 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr20_-_36793663 | 0.07 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr12_+_52643077 | 0.07 |
ENST00000553310.2
ENST00000544024.1 |
KRT86
|
keratin 86 |
| chr6_+_31620191 | 0.07 |
ENST00000375918.2
ENST00000375920.4 |
APOM
|
apolipoprotein M |
| chr20_-_36793774 | 0.07 |
ENST00000361475.2
|
TGM2
|
transglutaminase 2 |
| chr18_+_43914159 | 0.07 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr19_-_12941469 | 0.07 |
ENST00000586969.1
ENST00000589681.1 ENST00000585384.1 ENST00000589808.1 |
RTBDN
|
retbindin |
| chr6_+_144471643 | 0.07 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr15_-_60690932 | 0.07 |
ENST00000559818.1
|
ANXA2
|
annexin A2 |
| chr5_+_421030 | 0.07 |
ENST00000506456.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr17_-_41623716 | 0.07 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr14_+_51338816 | 0.06 |
ENST00000353130.1
ENST00000337334.2 ENST00000395752.1 |
ABHD12B
|
abhydrolase domain containing 12B |
| chr4_-_106817137 | 0.06 |
ENST00000510876.1
|
INTS12
|
integrator complex subunit 12 |
| chr22_-_24096630 | 0.06 |
ENST00000248948.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr16_-_66959429 | 0.06 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr11_-_125365435 | 0.06 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr2_-_188312971 | 0.06 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr20_-_61733657 | 0.06 |
ENST00000608031.1
ENST00000447910.2 |
HAR1B
|
highly accelerated region 1B (non-protein coding) |
| chr1_+_174769006 | 0.06 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr19_+_10563567 | 0.06 |
ENST00000344979.3
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr16_+_71660079 | 0.06 |
ENST00000565261.1
ENST00000268485.3 ENST00000299952.4 |
MARVELD3
|
MARVEL domain containing 3 |
| chr19_+_45973120 | 0.06 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr19_-_44405623 | 0.05 |
ENST00000591815.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr14_-_85996332 | 0.05 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr7_+_125078119 | 0.05 |
ENST00000458437.1
ENST00000415896.1 |
RP11-807H17.1
|
RP11-807H17.1 |
| chr17_+_5973740 | 0.05 |
ENST00000576083.1
|
WSCD1
|
WSC domain containing 1 |
| chr3_-_52868931 | 0.05 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr3_-_32022733 | 0.05 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chr11_-_64511575 | 0.05 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr6_-_31619742 | 0.05 |
ENST00000433828.1
ENST00000456286.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr2_-_236692031 | 0.05 |
ENST00000440101.1
|
AC064874.1
|
Uncharacterized protein |
| chr14_-_54420133 | 0.05 |
ENST00000559501.1
ENST00000558984.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr17_-_41623075 | 0.05 |
ENST00000545089.1
|
ETV4
|
ets variant 4 |
| chr3_-_168865522 | 0.05 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr8_-_11996586 | 0.04 |
ENST00000333796.3
|
USP17L2
|
ubiquitin specific peptidase 17-like family member 2 |
| chr6_-_90121938 | 0.04 |
ENST00000369415.4
|
RRAGD
|
Ras-related GTP binding D |
| chr1_+_56880606 | 0.04 |
ENST00000451914.1
|
RP4-710M16.2
|
RP4-710M16.2 |
| chr11_+_60869867 | 0.04 |
ENST00000347785.3
|
CD5
|
CD5 molecule |
| chr22_-_37545972 | 0.04 |
ENST00000216223.5
|
IL2RB
|
interleukin 2 receptor, beta |
| chr6_+_12012536 | 0.04 |
ENST00000379388.2
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr19_+_13051206 | 0.04 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr12_+_52668394 | 0.04 |
ENST00000423955.2
|
KRT86
|
keratin 86 |
| chr1_+_153651078 | 0.04 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr20_+_51588873 | 0.04 |
ENST00000371497.5
|
TSHZ2
|
teashirt zinc finger homeobox 2 |
| chr12_+_56414851 | 0.04 |
ENST00000547167.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr7_+_74188309 | 0.04 |
ENST00000289473.4
ENST00000433458.1 |
NCF1
|
neutrophil cytosolic factor 1 |
| chr6_-_90121789 | 0.04 |
ENST00000359203.3
|
RRAGD
|
Ras-related GTP binding D |
| chr16_+_89228757 | 0.04 |
ENST00000565008.1
|
LINC00304
|
long intergenic non-protein coding RNA 304 |
| chr22_+_42372764 | 0.04 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr14_+_45366518 | 0.04 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr19_-_43382142 | 0.04 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr8_-_102803163 | 0.04 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr1_+_116654376 | 0.04 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr7_-_27142290 | 0.04 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr19_+_30863271 | 0.04 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr3_+_52009110 | 0.04 |
ENST00000491470.1
|
ABHD14A
|
abhydrolase domain containing 14A |
| chr17_-_41623259 | 0.04 |
ENST00000538265.1
ENST00000591713.1 |
ETV4
|
ets variant 4 |
| chr1_-_171621815 | 0.04 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr5_+_122424816 | 0.04 |
ENST00000407847.4
|
PRDM6
|
PR domain containing 6 |
| chrX_-_30871004 | 0.03 |
ENST00000378928.1
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr15_-_30686052 | 0.03 |
ENST00000562729.1
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr1_-_98511756 | 0.03 |
ENST00000602984.1
ENST00000602852.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr19_+_37407212 | 0.03 |
ENST00000427117.1
ENST00000587130.1 ENST00000333987.7 ENST00000415168.1 ENST00000444991.1 |
ZNF568
|
zinc finger protein 568 |
| chr7_+_87257701 | 0.03 |
ENST00000338056.3
ENST00000493037.1 |
RUNDC3B
|
RUN domain containing 3B |
| chr19_-_43099070 | 0.03 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr17_-_75878647 | 0.03 |
ENST00000374983.2
|
FLJ45079
|
FLJ45079 |
| chr3_-_99833333 | 0.03 |
ENST00000354552.3
ENST00000331335.5 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr1_+_41249539 | 0.03 |
ENST00000347132.5
ENST00000509682.2 |
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chr7_+_73442457 | 0.03 |
ENST00000438880.1
ENST00000414324.1 ENST00000380562.4 |
ELN
|
elastin |
| chr16_+_57680811 | 0.03 |
ENST00000569101.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr1_-_72748417 | 0.03 |
ENST00000357731.5
|
NEGR1
|
neuronal growth regulator 1 |
| chr17_+_1182948 | 0.03 |
ENST00000333813.3
|
TUSC5
|
tumor suppressor candidate 5 |
| chr16_-_30022735 | 0.03 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr1_-_149889382 | 0.03 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chr11_-_119066545 | 0.03 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr15_-_53082178 | 0.03 |
ENST00000305901.5
|
ONECUT1
|
one cut homeobox 1 |
| chr19_+_30719410 | 0.03 |
ENST00000585628.1
ENST00000591488.1 |
ZNF536
|
zinc finger protein 536 |
| chr5_+_6448736 | 0.03 |
ENST00000399816.3
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr3_-_18466787 | 0.03 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr14_+_53019993 | 0.03 |
ENST00000542169.2
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr18_-_46784778 | 0.03 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr3_+_32737027 | 0.03 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr5_-_149669192 | 0.03 |
ENST00000398376.3
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr7_+_73442422 | 0.03 |
ENST00000358929.4
ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN
|
elastin |
| chr16_-_1031259 | 0.03 |
ENST00000563837.1
ENST00000563863.1 ENST00000565069.1 ENST00000570014.1 |
RP11-161M6.2
LMF1
|
RP11-161M6.2 lipase maturation factor 1 |
| chr15_+_96875657 | 0.03 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_+_56414795 | 0.03 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr2_-_74735707 | 0.03 |
ENST00000233630.6
|
PCGF1
|
polycomb group ring finger 1 |
| chr15_+_84841242 | 0.03 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr8_-_69243711 | 0.03 |
ENST00000512294.3
|
RP11-664D7.4
|
HCG1787533; Uncharacterized protein |
| chr6_+_97372645 | 0.03 |
ENST00000536676.1
ENST00000544166.1 |
KLHL32
|
kelch-like family member 32 |
| chr12_+_50451462 | 0.03 |
ENST00000447966.2
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr2_+_201170999 | 0.03 |
ENST00000439395.1
ENST00000444012.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr3_-_142607740 | 0.03 |
ENST00000485766.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr18_-_29340827 | 0.03 |
ENST00000269205.5
|
SLC25A52
|
solute carrier family 25, member 52 |
| chr11_-_69867159 | 0.03 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chr19_-_33716750 | 0.03 |
ENST00000253188.4
|
SLC7A10
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
| chr8_-_143696833 | 0.03 |
ENST00000356613.2
|
ARC
|
activity-regulated cytoskeleton-associated protein |
| chr4_-_5894777 | 0.03 |
ENST00000324989.7
|
CRMP1
|
collapsin response mediator protein 1 |
| chr7_+_114055052 | 0.03 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr19_+_50433476 | 0.03 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr2_+_220436917 | 0.03 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr1_+_113217345 | 0.03 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr17_+_72428266 | 0.03 |
ENST00000582473.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_+_123430948 | 0.03 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chrX_-_118739835 | 0.03 |
ENST00000542113.1
ENST00000304449.5 |
NKRF
|
NFKB repressing factor |
| chr19_-_52551814 | 0.03 |
ENST00000594154.1
ENST00000598745.1 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr11_-_61348292 | 0.03 |
ENST00000539008.1
ENST00000540677.1 ENST00000542836.1 ENST00000542670.1 ENST00000535826.1 ENST00000545053.1 |
SYT7
|
synaptotagmin VII |
| chr19_+_15619299 | 0.03 |
ENST00000269703.3
|
CYP4F22
|
cytochrome P450, family 4, subfamily F, polypeptide 22 |
| chrX_-_119445306 | 0.03 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr11_+_8932654 | 0.02 |
ENST00000299576.5
ENST00000309377.4 ENST00000309357.4 |
AKIP1
|
A kinase (PRKA) interacting protein 1 |
| chr1_+_218683438 | 0.02 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr7_+_87257854 | 0.02 |
ENST00000394654.3
|
RUNDC3B
|
RUN domain containing 3B |
| chr5_-_158757895 | 0.02 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr19_-_10227503 | 0.02 |
ENST00000593054.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr5_+_137225125 | 0.02 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr7_+_27282319 | 0.02 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr20_+_36888551 | 0.02 |
ENST00000418004.1
ENST00000451435.1 |
BPI
|
bactericidal/permeability-increasing protein |
| chr1_+_247712383 | 0.02 |
ENST00000366488.4
ENST00000536561.1 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chr16_+_2564254 | 0.02 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr10_+_99609996 | 0.02 |
ENST00000370602.1
|
GOLGA7B
|
golgin A7 family, member B |
| chr11_+_7598239 | 0.02 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr5_-_1445539 | 0.02 |
ENST00000270349.9
|
SLC6A3
|
solute carrier family 6 (neurotransmitter transporter), member 3 |
| chr12_+_107349606 | 0.02 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr8_+_21914477 | 0.02 |
ENST00000517804.1
|
DMTN
|
dematin actin binding protein |
| chr2_-_56150184 | 0.02 |
ENST00000394554.1
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr7_-_100823496 | 0.02 |
ENST00000455377.1
ENST00000443096.1 ENST00000300303.2 |
NAT16
|
N-acetyltransferase 16 (GCN5-related, putative) |
| chr4_-_70626430 | 0.02 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr6_-_31620455 | 0.02 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chr4_+_144258288 | 0.02 |
ENST00000514639.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr19_-_55691614 | 0.02 |
ENST00000592470.1
ENST00000354308.3 |
SYT5
|
synaptotagmin V |
| chr8_+_145064233 | 0.02 |
ENST00000529301.1
ENST00000395068.4 |
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr11_+_308217 | 0.02 |
ENST00000602569.1
|
IFITM2
|
interferon induced transmembrane protein 2 |
| chr19_-_42806842 | 0.02 |
ENST00000596265.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr12_+_50451331 | 0.02 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr3_-_196065374 | 0.02 |
ENST00000454715.1
|
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr12_-_49259643 | 0.02 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chrX_-_119445263 | 0.02 |
ENST00000309720.5
|
TMEM255A
|
transmembrane protein 255A |
| chr10_+_104154229 | 0.02 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr12_+_107349497 | 0.02 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr12_+_56473910 | 0.02 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr11_-_89224638 | 0.02 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr1_+_113217073 | 0.02 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr19_+_18530146 | 0.02 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chrX_+_100474763 | 0.02 |
ENST00000395209.3
|
DRP2
|
dystrophin related protein 2 |
| chr20_-_42816206 | 0.02 |
ENST00000372980.3
|
JPH2
|
junctophilin 2 |
| chr12_-_109025849 | 0.02 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr2_-_197226875 | 0.02 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_46668994 | 0.02 |
ENST00000371980.3
|
LURAP1
|
leucine rich adaptor protein 1 |
| chr4_+_40198527 | 0.02 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr1_+_113217043 | 0.02 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr4_+_81951957 | 0.02 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr11_+_66512303 | 0.02 |
ENST00000532565.2
|
C11orf80
|
chromosome 11 open reading frame 80 |
| chr19_-_43709772 | 0.02 |
ENST00000596907.1
ENST00000451895.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr19_-_37958086 | 0.02 |
ENST00000592490.1
ENST00000392149.2 |
ZNF569
|
zinc finger protein 569 |
| chr13_+_26828275 | 0.02 |
ENST00000381527.3
|
CDK8
|
cyclin-dependent kinase 8 |
| chr12_-_26278030 | 0.02 |
ENST00000242728.4
|
BHLHE41
|
basic helix-loop-helix family, member e41 |
| chr3_-_18466026 | 0.02 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr1_+_160336851 | 0.02 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr4_+_113066552 | 0.02 |
ENST00000309733.5
|
C4orf32
|
chromosome 4 open reading frame 32 |
| chr21_-_43786634 | 0.02 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr21_+_34619079 | 0.02 |
ENST00000433395.2
|
AP000295.9
|
AP000295.9 |
| chr15_+_41245160 | 0.02 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr2_+_112895939 | 0.02 |
ENST00000331203.2
ENST00000409903.1 ENST00000409667.3 ENST00000409450.3 |
FBLN7
|
fibulin 7 |
| chr11_-_441964 | 0.02 |
ENST00000332826.6
|
ANO9
|
anoctamin 9 |
| chr1_-_201390846 | 0.02 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr22_+_41968007 | 0.02 |
ENST00000460790.1
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr16_+_57680840 | 0.02 |
ENST00000563862.1
ENST00000564722.1 ENST00000569158.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr12_+_133707204 | 0.02 |
ENST00000426665.2
|
ZNF10
|
zinc finger protein 10 |
| chr22_-_39637135 | 0.02 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr1_+_113217309 | 0.02 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr19_-_55691472 | 0.02 |
ENST00000537500.1
|
SYT5
|
synaptotagmin V |
| chr10_+_127661942 | 0.02 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr17_+_78194261 | 0.02 |
ENST00000572725.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr11_-_22647350 | 0.02 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr9_+_34653861 | 0.02 |
ENST00000556792.1
ENST00000318041.9 ENST00000378817.4 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr13_+_88325498 | 0.02 |
ENST00000400028.3
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EPAS1_BCL3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0044115 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0035284 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |