Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
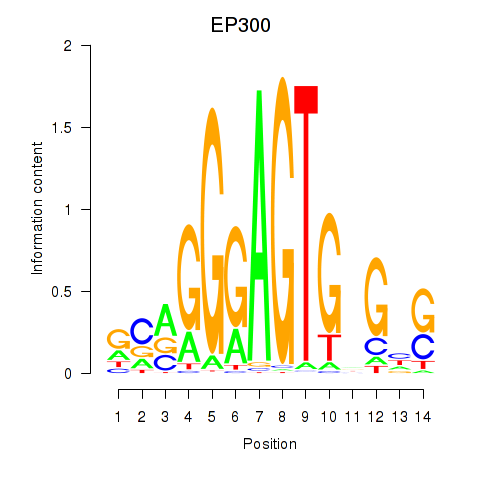
Results for EP300
Z-value: 0.62
Transcription factors associated with EP300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EP300
|
ENSG00000100393.9 | E1A binding protein p300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EP300 | hg19_v2_chr22_+_41487711_41487798 | 0.41 | 5.9e-01 | Click! |
Activity profile of EP300 motif
Sorted Z-values of EP300 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_28443819 | 0.34 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr11_-_64527425 | 0.24 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr2_+_217277271 | 0.22 |
ENST00000425815.1
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr2_+_20866424 | 0.21 |
ENST00000272224.3
|
GDF7
|
growth differentiation factor 7 |
| chr12_+_7072354 | 0.20 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr1_-_153513170 | 0.19 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr10_+_81892477 | 0.18 |
ENST00000372263.3
|
PLAC9
|
placenta-specific 9 |
| chr12_+_72058130 | 0.17 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr11_+_46403303 | 0.16 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr6_+_20403997 | 0.16 |
ENST00000535432.1
|
E2F3
|
E2F transcription factor 3 |
| chr11_+_46403194 | 0.16 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr4_+_184427235 | 0.15 |
ENST00000412117.1
ENST00000434682.2 |
ING2
|
inhibitor of growth family, member 2 |
| chr1_+_153651078 | 0.15 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr2_+_110371905 | 0.15 |
ENST00000356454.3
|
SOWAHC
|
sosondowah ankyrin repeat domain family member C |
| chr14_+_56584414 | 0.15 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr19_-_11545920 | 0.14 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr11_+_46722368 | 0.14 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr11_+_46402583 | 0.14 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr20_+_54987305 | 0.13 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr8_-_37594944 | 0.13 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr14_+_77582905 | 0.13 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr11_+_46402744 | 0.13 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr5_-_443239 | 0.13 |
ENST00000408966.2
|
C5orf55
|
chromosome 5 open reading frame 55 |
| chr8_-_33424636 | 0.13 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chr1_+_211433275 | 0.13 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr19_-_56109119 | 0.12 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr7_-_944631 | 0.12 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr6_-_30684898 | 0.12 |
ENST00000422266.1
ENST00000416571.1 |
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr1_-_11865982 | 0.12 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr20_+_54987168 | 0.12 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr4_-_99064387 | 0.12 |
ENST00000295268.3
|
STPG2
|
sperm-tail PG-rich repeat containing 2 |
| chr7_-_5569588 | 0.11 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr13_-_44361025 | 0.11 |
ENST00000261488.6
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr3_-_126076264 | 0.11 |
ENST00000296233.3
|
KLF15
|
Kruppel-like factor 15 |
| chr12_-_81331460 | 0.11 |
ENST00000549417.1
|
LIN7A
|
lin-7 homolog A (C. elegans) |
| chr8_+_144816303 | 0.11 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr3_-_32022733 | 0.10 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chrX_-_45710920 | 0.10 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr1_-_43855444 | 0.10 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr7_+_108210012 | 0.10 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr6_-_28226984 | 0.10 |
ENST00000423974.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr11_-_2924720 | 0.10 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr2_+_87808725 | 0.10 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr1_+_6845578 | 0.10 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr3_-_10028366 | 0.10 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chrX_-_140271249 | 0.10 |
ENST00000370526.2
|
LDOC1
|
leucine zipper, down-regulated in cancer 1 |
| chr17_-_79995553 | 0.10 |
ENST00000581584.1
ENST00000577712.1 ENST00000582900.1 ENST00000579155.1 ENST00000306869.2 |
DCXR
|
dicarbonyl/L-xylulose reductase |
| chr19_-_18649181 | 0.09 |
ENST00000596015.1
|
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr1_+_32827759 | 0.09 |
ENST00000373534.3
|
TSSK3
|
testis-specific serine kinase 3 |
| chr16_-_29465668 | 0.09 |
ENST00000569622.1
|
RP11-345J4.5
|
BolA-like protein 2 |
| chr1_-_209957882 | 0.09 |
ENST00000294811.1
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr12_-_109027643 | 0.09 |
ENST00000388962.3
ENST00000550948.1 |
SELPLG
|
selectin P ligand |
| chr15_+_74466012 | 0.09 |
ENST00000249842.3
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr1_-_155658260 | 0.09 |
ENST00000368339.5
ENST00000405763.3 ENST00000368340.5 ENST00000454523.1 ENST00000443231.1 ENST00000347088.5 ENST00000361831.5 ENST00000355499.4 |
YY1AP1
|
YY1 associated protein 1 |
| chr19_-_56988677 | 0.09 |
ENST00000504904.3
ENST00000292069.6 |
ZNF667
|
zinc finger protein 667 |
| chr2_-_70417827 | 0.09 |
ENST00000457952.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr16_-_30022735 | 0.09 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr1_+_116519223 | 0.08 |
ENST00000369502.1
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr19_-_6110555 | 0.08 |
ENST00000593241.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr6_+_43738444 | 0.08 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr2_+_74153953 | 0.08 |
ENST00000264093.4
ENST00000348222.1 |
DGUOK
|
deoxyguanosine kinase |
| chr12_+_116997186 | 0.08 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr2_-_110371720 | 0.08 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr15_-_93353028 | 0.08 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr20_+_10199468 | 0.08 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr11_-_118122996 | 0.08 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr22_-_31688381 | 0.08 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_-_149093499 | 0.08 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr5_+_137225125 | 0.08 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr2_+_201170999 | 0.08 |
ENST00000439395.1
ENST00000444012.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr22_+_20105259 | 0.08 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr19_+_843314 | 0.08 |
ENST00000544537.2
|
PRTN3
|
proteinase 3 |
| chr1_+_6845497 | 0.08 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr7_-_99149715 | 0.08 |
ENST00000449309.1
|
FAM200A
|
family with sequence similarity 200, member A |
| chr1_-_95007193 | 0.08 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr3_-_130745571 | 0.08 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr8_-_23021533 | 0.08 |
ENST00000312584.3
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
| chr12_-_48213735 | 0.08 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr16_+_84682108 | 0.08 |
ENST00000564996.1
ENST00000258157.5 ENST00000567410.1 |
KLHL36
|
kelch-like family member 36 |
| chr17_-_7080227 | 0.07 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr15_-_74726283 | 0.07 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr1_+_95582881 | 0.07 |
ENST00000370203.4
ENST00000456991.1 |
TMEM56
|
transmembrane protein 56 |
| chr11_+_62623512 | 0.07 |
ENST00000377892.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr11_+_62623544 | 0.07 |
ENST00000377890.2
ENST00000377891.2 ENST00000377889.2 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr1_+_204485503 | 0.07 |
ENST00000367182.3
ENST00000507825.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr12_-_129308041 | 0.07 |
ENST00000376740.4
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr2_-_110371777 | 0.07 |
ENST00000397712.2
|
SEPT10
|
septin 10 |
| chrX_+_91034260 | 0.07 |
ENST00000395337.2
|
PCDH11X
|
protocadherin 11 X-linked |
| chr5_+_139028510 | 0.07 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr14_-_65346555 | 0.07 |
ENST00000542895.1
ENST00000556626.1 |
SPTB
|
spectrin, beta, erythrocytic |
| chr1_-_1051736 | 0.07 |
ENST00000448924.1
ENST00000294576.5 ENST00000437760.1 ENST00000462097.1 ENST00000475119.1 |
C1orf159
|
chromosome 1 open reading frame 159 |
| chrX_-_107018969 | 0.07 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr16_-_30204987 | 0.07 |
ENST00000569282.1
ENST00000567436.1 |
BOLA2B
|
bolA family member 2B |
| chr1_-_43855479 | 0.07 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr11_+_102188272 | 0.07 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr1_+_116519112 | 0.07 |
ENST00000369503.4
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr7_-_5570229 | 0.07 |
ENST00000331789.5
|
ACTB
|
actin, beta |
| chr11_+_3875757 | 0.07 |
ENST00000525403.1
|
STIM1
|
stromal interaction molecule 1 |
| chr16_-_27279900 | 0.07 |
ENST00000564342.1
ENST00000567710.1 ENST00000563273.1 ENST00000361439.4 |
NSMCE1
|
non-SMC element 1 homolog (S. cerevisiae) |
| chr6_+_3000195 | 0.07 |
ENST00000338130.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr9_-_123638633 | 0.07 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr16_+_69458428 | 0.06 |
ENST00000512062.1
ENST00000307892.8 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr20_+_61340179 | 0.06 |
ENST00000370501.3
|
NTSR1
|
neurotensin receptor 1 (high affinity) |
| chr2_+_217277466 | 0.06 |
ENST00000358207.5
ENST00000434435.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr11_-_46722117 | 0.06 |
ENST00000311956.4
|
ARHGAP1
|
Rho GTPase activating protein 1 |
| chr8_+_95732095 | 0.06 |
ENST00000414645.2
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr19_-_11039188 | 0.06 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr11_-_126081532 | 0.06 |
ENST00000533628.1
ENST00000298317.4 ENST00000532674.1 |
RPUSD4
|
RNA pseudouridylate synthase domain containing 4 |
| chr17_+_79031415 | 0.06 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr17_-_56065540 | 0.06 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr6_+_3000218 | 0.06 |
ENST00000380441.1
ENST00000380455.4 ENST00000380454.4 |
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr14_-_69445793 | 0.06 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr15_-_73925651 | 0.06 |
ENST00000545878.1
ENST00000287226.8 ENST00000345330.4 |
NPTN
|
neuroplastin |
| chr10_-_69835001 | 0.06 |
ENST00000513996.1
ENST00000412272.2 ENST00000395198.3 ENST00000492996.2 |
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr11_+_63870660 | 0.06 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr12_-_49393092 | 0.06 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr11_+_236959 | 0.06 |
ENST00000431206.2
ENST00000528906.1 |
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr19_+_11909329 | 0.06 |
ENST00000323169.5
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr1_-_157108266 | 0.06 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr1_+_155657737 | 0.06 |
ENST00000471642.2
ENST00000471214.1 |
DAP3
|
death associated protein 3 |
| chr12_-_110888103 | 0.06 |
ENST00000426440.1
ENST00000228825.7 |
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr22_+_20105012 | 0.06 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr19_-_49955096 | 0.06 |
ENST00000595550.1
|
PIH1D1
|
PIH1 domain containing 1 |
| chr5_+_137225158 | 0.06 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr1_-_161102367 | 0.06 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr11_+_102188224 | 0.06 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr5_-_1345199 | 0.06 |
ENST00000320895.5
|
CLPTM1L
|
CLPTM1-like |
| chr15_-_52861394 | 0.06 |
ENST00000563277.1
ENST00000566423.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr10_-_99161033 | 0.06 |
ENST00000315563.6
ENST00000370992.4 ENST00000414986.1 |
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr6_-_33290580 | 0.06 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr2_-_110371412 | 0.06 |
ENST00000415095.1
ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10
|
septin 10 |
| chr1_-_84543614 | 0.06 |
ENST00000605506.1
|
RP11-486G15.2
|
RP11-486G15.2 |
| chr11_+_62623621 | 0.06 |
ENST00000535296.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr19_+_49617609 | 0.06 |
ENST00000221459.2
ENST00000486217.2 |
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr8_-_103668114 | 0.06 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr12_-_57119068 | 0.06 |
ENST00000546862.1
ENST00000550952.1 ENST00000548563.1 |
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr2_+_30369859 | 0.06 |
ENST00000402003.3
|
YPEL5
|
yippee-like 5 (Drosophila) |
| chr16_-_28222797 | 0.06 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr2_-_70418032 | 0.06 |
ENST00000425268.1
ENST00000428751.1 ENST00000417203.1 ENST00000417865.1 ENST00000428010.1 ENST00000447804.1 ENST00000264434.2 |
C2orf42
|
chromosome 2 open reading frame 42 |
| chr10_-_16859361 | 0.06 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr11_-_3663480 | 0.06 |
ENST00000397068.3
|
ART5
|
ADP-ribosyltransferase 5 |
| chr11_-_3862059 | 0.06 |
ENST00000396978.1
|
RHOG
|
ras homolog family member G |
| chr2_+_74154032 | 0.06 |
ENST00000356837.6
|
DGUOK
|
deoxyguanosine kinase |
| chr12_-_57119300 | 0.06 |
ENST00000546917.1
ENST00000454682.1 |
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr14_+_23564700 | 0.05 |
ENST00000554203.1
|
C14orf119
|
chromosome 14 open reading frame 119 |
| chr3_-_172428959 | 0.05 |
ENST00000475381.1
ENST00000538775.1 ENST00000273512.3 ENST00000543711.1 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr18_+_3262415 | 0.05 |
ENST00000581193.1
ENST00000400175.5 |
MYL12B
|
myosin, light chain 12B, regulatory |
| chr5_-_1344999 | 0.05 |
ENST00000320927.6
|
CLPTM1L
|
CLPTM1-like |
| chr20_+_55926583 | 0.05 |
ENST00000395840.2
|
RAE1
|
ribonucleic acid export 1 |
| chr11_+_3876859 | 0.05 |
ENST00000300737.4
|
STIM1
|
stromal interaction molecule 1 |
| chr2_+_217277137 | 0.05 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr13_+_51796497 | 0.05 |
ENST00000322475.8
ENST00000280057.6 |
FAM124A
|
family with sequence similarity 124A |
| chr6_+_2988199 | 0.05 |
ENST00000450238.1
ENST00000445000.1 ENST00000426637.1 |
LINC01011
NQO2
|
long intergenic non-protein coding RNA 1011 NAD(P)H dehydrogenase, quinone 2 |
| chr2_-_241075706 | 0.05 |
ENST00000607357.1
ENST00000307266.3 |
MYEOV2
|
myeloma overexpressed 2 |
| chr10_-_95360983 | 0.05 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr15_-_52861029 | 0.05 |
ENST00000561650.1
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr12_-_49525175 | 0.05 |
ENST00000336023.5
ENST00000550367.1 ENST00000552984.1 ENST00000547476.1 |
TUBA1B
|
tubulin, alpha 1b |
| chr22_+_46731676 | 0.05 |
ENST00000424260.2
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr19_-_17186229 | 0.05 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr14_+_52327109 | 0.05 |
ENST00000335281.4
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chrX_-_107019181 | 0.05 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chrX_-_131352152 | 0.05 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr9_+_140149625 | 0.05 |
ENST00000343053.4
|
NELFB
|
negative elongation factor complex member B |
| chr17_+_7477040 | 0.05 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr15_+_38544476 | 0.05 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr8_-_91657909 | 0.05 |
ENST00000418210.2
|
TMEM64
|
transmembrane protein 64 |
| chr19_+_10362882 | 0.05 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr1_-_2126174 | 0.05 |
ENST00000400919.3
ENST00000420515.1 ENST00000378543.2 ENST00000400918.3 |
C1orf86
|
chromosome 1 open reading frame 86 |
| chr5_+_175085033 | 0.05 |
ENST00000377291.2
|
HRH2
|
histamine receptor H2 |
| chr16_-_69373396 | 0.05 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr11_+_2421718 | 0.05 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr19_+_29704142 | 0.05 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr12_-_105630016 | 0.05 |
ENST00000258530.3
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr6_-_27440837 | 0.05 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr19_-_49955050 | 0.05 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chr11_+_105481405 | 0.05 |
ENST00000428631.2
|
GRIA4
|
glutamate receptor, ionotropic, AMPA 4 |
| chr18_+_9334755 | 0.05 |
ENST00000262120.5
|
TWSG1
|
twisted gastrulation BMP signaling modulator 1 |
| chr3_+_184080790 | 0.05 |
ENST00000430783.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_+_237016 | 0.05 |
ENST00000352303.5
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr18_+_32556892 | 0.05 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_-_155658085 | 0.05 |
ENST00000311573.5
ENST00000438245.2 |
YY1AP1
|
YY1 associated protein 1 |
| chr1_-_25256368 | 0.05 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chrX_-_48326683 | 0.05 |
ENST00000440085.1
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr20_-_30311703 | 0.05 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr1_-_155658518 | 0.05 |
ENST00000404643.1
ENST00000359205.5 ENST00000407221.1 |
YY1AP1
|
YY1 associated protein 1 |
| chr16_+_2083265 | 0.05 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr16_-_66968055 | 0.05 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr6_-_27440460 | 0.05 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr12_+_122241928 | 0.05 |
ENST00000604567.1
ENST00000542440.1 |
SETD1B
|
SET domain containing 1B |
| chr14_-_36990354 | 0.05 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr3_-_135914615 | 0.05 |
ENST00000309993.2
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr22_-_20104700 | 0.05 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr9_-_33473882 | 0.05 |
ENST00000455041.2
ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6
|
nucleolar protein 6 (RNA-associated) |
| chr6_-_24720226 | 0.05 |
ENST00000378102.3
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr11_+_236540 | 0.05 |
ENST00000532097.1
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr6_-_35888905 | 0.05 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr6_-_110500905 | 0.04 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr6_-_35888824 | 0.04 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr19_+_46195895 | 0.04 |
ENST00000366382.4
|
QPCTL
|
glutaminyl-peptide cyclotransferase-like |
| chr16_+_76311169 | 0.04 |
ENST00000307431.8
ENST00000377504.4 |
CNTNAP4
|
contactin associated protein-like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EP300
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.2 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.0 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.0 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:1902683 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.0 | GO:0006218 | uridine catabolic process(GO:0006218) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.0 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |