Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
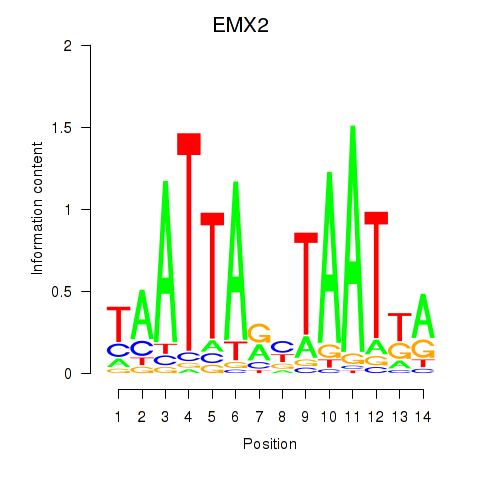
Results for EMX2
Z-value: 0.35
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.10 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg19_v2_chr10_+_119302508_119302559 | 0.87 | 1.3e-01 | Click! |
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26235206 | 0.40 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr4_+_174818390 | 0.16 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr5_+_42756903 | 0.14 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr1_+_149754227 | 0.13 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr16_+_15489629 | 0.13 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr20_-_29896388 | 0.13 |
ENST00000400549.1
|
DEFB116
|
defensin, beta 116 |
| chr16_+_15489603 | 0.13 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr5_+_136070614 | 0.12 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr4_-_112993808 | 0.12 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr2_-_207082748 | 0.12 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr2_-_236684438 | 0.11 |
ENST00000409661.1
|
AC064874.1
|
Uncharacterized protein |
| chr2_-_99279928 | 0.10 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr11_+_122709200 | 0.10 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr6_+_26204825 | 0.10 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr6_+_27833034 | 0.10 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr15_+_75970672 | 0.09 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr19_-_35264089 | 0.09 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chr12_+_78359999 | 0.09 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chrX_+_47444613 | 0.09 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr17_-_4938712 | 0.08 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr2_-_99871570 | 0.08 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr16_+_28763108 | 0.08 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr15_+_63188009 | 0.08 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr1_+_159931002 | 0.08 |
ENST00000443364.1
ENST00000423943.1 |
RP11-48O20.4
|
long intergenic non-protein coding RNA 1133 |
| chr5_-_140013224 | 0.08 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr17_-_57158523 | 0.07 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr12_-_100656134 | 0.07 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr15_-_99057551 | 0.07 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr3_-_58523010 | 0.07 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr1_+_218683438 | 0.07 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr6_-_86099898 | 0.07 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr8_+_101349823 | 0.07 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr2_-_14541060 | 0.07 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr12_+_28605426 | 0.07 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr5_-_150727111 | 0.07 |
ENST00000335244.4
ENST00000521967.1 |
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr2_+_39117010 | 0.06 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr3_+_140981456 | 0.06 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr4_-_185275104 | 0.06 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr3_-_20053741 | 0.06 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr8_-_38008783 | 0.06 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr2_+_105050794 | 0.06 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr14_-_20922960 | 0.06 |
ENST00000553640.1
ENST00000488532.2 |
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr12_-_110883346 | 0.06 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr21_-_27423339 | 0.06 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr7_+_13141010 | 0.06 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr16_+_28648975 | 0.06 |
ENST00000529716.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr19_-_58864848 | 0.06 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr11_-_124981475 | 0.06 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr12_+_72061563 | 0.06 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr6_-_138539627 | 0.06 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr19_-_51920952 | 0.06 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr19_-_43969796 | 0.05 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr6_+_63921351 | 0.05 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr12_-_123201337 | 0.05 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr12_-_49581152 | 0.05 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr16_-_28374829 | 0.05 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr18_-_46895066 | 0.05 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr2_-_74007095 | 0.05 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr5_-_140013275 | 0.05 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr17_+_7239821 | 0.05 |
ENST00000158762.3
ENST00000570457.2 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr3_-_130745571 | 0.05 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr20_+_57414743 | 0.05 |
ENST00000313949.7
|
GNAS
|
GNAS complex locus |
| chr11_+_12308447 | 0.05 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr17_+_79849872 | 0.05 |
ENST00000584197.1
ENST00000583839.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr12_+_94071129 | 0.05 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr9_-_128246769 | 0.05 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr10_-_79397740 | 0.05 |
ENST00000372440.1
ENST00000480683.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr9_+_93759317 | 0.05 |
ENST00000563268.1
|
RP11-367F23.2
|
RP11-367F23.2 |
| chr3_-_127455200 | 0.05 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr11_-_62609281 | 0.05 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr1_+_192778161 | 0.05 |
ENST00000235382.5
|
RGS2
|
regulator of G-protein signaling 2, 24kDa |
| chr11_+_92085262 | 0.05 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr18_+_616672 | 0.05 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr6_+_13272904 | 0.05 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr10_-_74283694 | 0.05 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr2_+_161993465 | 0.05 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr22_-_40929812 | 0.05 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr11_-_114430570 | 0.04 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr12_-_112279694 | 0.04 |
ENST00000443596.1
ENST00000442119.1 |
MAPKAPK5-AS1
|
MAPKAPK5 antisense RNA 1 |
| chr3_+_63638372 | 0.04 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr2_-_85625857 | 0.04 |
ENST00000453973.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr3_+_52245458 | 0.04 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr3_-_48956818 | 0.04 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr4_-_89205879 | 0.04 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr6_+_30585486 | 0.04 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr7_+_115862858 | 0.04 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr3_+_149191723 | 0.04 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr16_-_28634874 | 0.04 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr12_+_110011571 | 0.04 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr8_-_144660771 | 0.04 |
ENST00000449291.2
|
NAPRT1
|
nicotinate phosphoribosyltransferase domain containing 1 |
| chr12_-_10282742 | 0.04 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr1_-_139379 | 0.04 |
ENST00000423372.3
|
AL627309.1
|
Uncharacterized protein |
| chr6_-_134639180 | 0.04 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr12_+_94071341 | 0.04 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr3_+_113616317 | 0.04 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr1_-_67142710 | 0.04 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr17_-_39093672 | 0.04 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr2_+_196313239 | 0.04 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr10_-_71169031 | 0.04 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr19_-_36822551 | 0.04 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chrX_-_23926004 | 0.04 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr5_+_129083772 | 0.04 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr11_-_36310958 | 0.04 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr19_-_39340563 | 0.04 |
ENST00000601813.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr3_-_194188956 | 0.04 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr11_-_107729504 | 0.04 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr19_+_50016610 | 0.04 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr15_-_78913628 | 0.04 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr8_+_35649365 | 0.04 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr3_+_184018352 | 0.04 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr19_+_14693888 | 0.03 |
ENST00000547437.1
ENST00000397439.2 ENST00000417570.1 |
CLEC17A
|
C-type lectin domain family 17, member A |
| chr12_+_58176525 | 0.03 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr11_-_107729287 | 0.03 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr17_-_67138015 | 0.03 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr5_-_114938090 | 0.03 |
ENST00000427199.2
|
TICAM2
|
toll-like receptor adaptor molecule 2 |
| chr16_+_31885079 | 0.03 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr8_+_31497271 | 0.03 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr15_+_84115868 | 0.03 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr13_+_108921977 | 0.03 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr5_-_74162739 | 0.03 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr12_-_10282836 | 0.03 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr9_-_33473882 | 0.03 |
ENST00000455041.2
ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6
|
nucleolar protein 6 (RNA-associated) |
| chr4_-_170897045 | 0.03 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr17_+_42422662 | 0.03 |
ENST00000593167.1
ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN
|
granulin |
| chr14_+_31959154 | 0.03 |
ENST00000550005.1
|
NUBPL
|
nucleotide binding protein-like |
| chr7_-_111032971 | 0.03 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr18_+_61637159 | 0.03 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr5_+_81575281 | 0.03 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr10_-_75415825 | 0.03 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr1_-_110283660 | 0.03 |
ENST00000361066.2
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr21_+_17553910 | 0.03 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr11_-_62389621 | 0.03 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr1_+_56046710 | 0.03 |
ENST00000422374.1
|
RP11-466L17.1
|
RP11-466L17.1 |
| chr11_+_2405833 | 0.03 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chrX_+_77359671 | 0.03 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr12_+_80603233 | 0.03 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr18_+_21693306 | 0.03 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr1_+_10292308 | 0.03 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chr8_+_30244580 | 0.03 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr18_-_64271363 | 0.03 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr9_-_115095123 | 0.03 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr20_+_33563206 | 0.03 |
ENST00000262873.7
|
MYH7B
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr6_+_26251835 | 0.02 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr5_-_132200477 | 0.02 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr18_+_21572737 | 0.02 |
ENST00000304621.6
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr16_+_69599861 | 0.02 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr7_+_1727755 | 0.02 |
ENST00000424383.2
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr6_+_150690028 | 0.02 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr17_+_42422629 | 0.02 |
ENST00000589536.1
ENST00000587109.1 ENST00000587518.1 |
GRN
|
granulin |
| chr5_+_112312416 | 0.02 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr4_+_80584903 | 0.02 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr8_-_87242589 | 0.02 |
ENST00000419776.2
ENST00000297524.3 |
SLC7A13
|
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr1_+_146373546 | 0.02 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr8_-_40200877 | 0.02 |
ENST00000521030.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chrX_-_15402498 | 0.02 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr4_-_120243545 | 0.02 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr12_+_112279782 | 0.02 |
ENST00000550735.2
|
MAPKAPK5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr21_-_19858196 | 0.02 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr8_+_119294456 | 0.02 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr4_+_95128748 | 0.02 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr17_-_15244894 | 0.02 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr4_-_89205705 | 0.02 |
ENST00000295908.7
ENST00000510548.2 ENST00000508256.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr22_+_24407642 | 0.02 |
ENST00000454754.1
ENST00000263119.5 |
CABIN1
|
calcineurin binding protein 1 |
| chr5_+_118812294 | 0.02 |
ENST00000509514.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr8_+_24151553 | 0.02 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr3_-_109056419 | 0.02 |
ENST00000335658.6
|
DPPA4
|
developmental pluripotency associated 4 |
| chr12_+_25055243 | 0.02 |
ENST00000599478.1
|
AC026310.1
|
Protein 101060047 |
| chr12_+_26164645 | 0.02 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_-_119682812 | 0.02 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr21_+_17792672 | 0.02 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chrX_+_77359726 | 0.02 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr8_+_11961898 | 0.02 |
ENST00000400085.3
|
ZNF705D
|
zinc finger protein 705D |
| chr1_-_19615744 | 0.02 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr20_+_30697298 | 0.02 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr5_-_76788317 | 0.02 |
ENST00000296679.4
|
WDR41
|
WD repeat domain 41 |
| chr3_-_108248169 | 0.02 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr3_+_32993065 | 0.02 |
ENST00000330953.5
|
CCR4
|
chemokine (C-C motif) receptor 4 |
| chr17_-_4167142 | 0.02 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr12_-_58220078 | 0.02 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr17_-_42100474 | 0.02 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr12_+_29302119 | 0.02 |
ENST00000536681.3
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr17_+_42422637 | 0.02 |
ENST00000053867.3
ENST00000588143.1 |
GRN
|
granulin |
| chr1_-_86848760 | 0.02 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr2_-_179343226 | 0.02 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr14_+_62164340 | 0.02 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr18_+_29027696 | 0.02 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr10_+_122610687 | 0.02 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr6_+_42018614 | 0.02 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr5_+_118812237 | 0.02 |
ENST00000513628.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr13_-_30880979 | 0.02 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr22_+_50981079 | 0.02 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr4_-_186570679 | 0.02 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_55237484 | 0.02 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr12_-_106480587 | 0.02 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr7_+_107224364 | 0.02 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr16_+_29832634 | 0.02 |
ENST00000565164.1
ENST00000570234.1 |
MVP
|
major vault protein |
| chr1_+_212475148 | 0.02 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr14_-_69261310 | 0.02 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr17_-_60883993 | 0.02 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr3_+_111393501 | 0.02 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0017143 | insecticide metabolic process(GO:0017143) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.0 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.0 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |