Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
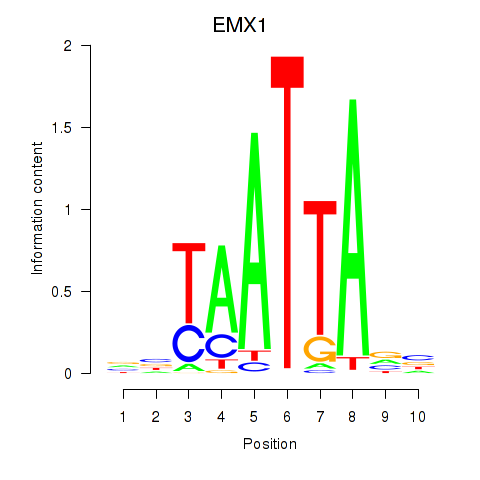
Results for EMX1
Z-value: 0.72
Transcription factors associated with EMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX1
|
ENSG00000135638.9 | empty spiracles homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX1 | hg19_v2_chr2_+_73144604_73144654 | 0.78 | 2.2e-01 | Click! |
Activity profile of EMX1 motif
Sorted Z-values of EMX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_75488984 | 0.61 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr13_+_110958124 | 0.55 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr17_+_61151306 | 0.50 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr8_+_94752349 | 0.44 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr16_+_53133070 | 0.40 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr8_-_25281747 | 0.39 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr16_+_12059091 | 0.37 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr8_-_1922789 | 0.35 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr1_+_225600404 | 0.31 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr2_-_203735586 | 0.30 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr6_-_32157947 | 0.29 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr9_-_5304432 | 0.29 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr5_+_148737562 | 0.28 |
ENST00000274569.4
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chrX_-_73512177 | 0.27 |
ENST00000603672.1
ENST00000418855.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr14_+_20187174 | 0.26 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chrX_-_77150911 | 0.26 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr9_-_117267717 | 0.25 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chrX_+_102024075 | 0.24 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr17_-_71223839 | 0.24 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr8_+_22424551 | 0.23 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr2_-_27498208 | 0.23 |
ENST00000424577.1
ENST00000426569.1 |
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr5_-_137374288 | 0.23 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr15_-_55562451 | 0.23 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr7_-_92777606 | 0.23 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr11_-_60010556 | 0.23 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr2_-_190446738 | 0.22 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr11_-_96076334 | 0.22 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr11_-_111649015 | 0.22 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr7_-_111424506 | 0.22 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr8_+_32579271 | 0.22 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr12_-_91546926 | 0.21 |
ENST00000550758.1
|
DCN
|
decorin |
| chr7_-_86849836 | 0.21 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr2_-_216003127 | 0.21 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr11_+_77532233 | 0.20 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr11_-_111649074 | 0.20 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr7_+_23719749 | 0.20 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr12_-_46121554 | 0.20 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chrX_-_118986911 | 0.19 |
ENST00000276201.2
ENST00000345865.2 |
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr7_-_111424462 | 0.19 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr14_+_61654271 | 0.19 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr4_-_69111401 | 0.19 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr4_+_169633310 | 0.18 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr8_-_19614810 | 0.17 |
ENST00000524213.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_+_32579321 | 0.17 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr14_-_51027838 | 0.17 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr19_-_52307357 | 0.17 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr2_+_149402989 | 0.17 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr8_-_29120604 | 0.16 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr1_-_21625486 | 0.16 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr12_-_10978957 | 0.16 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr10_-_21186144 | 0.16 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr12_-_118796971 | 0.15 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr19_-_12889226 | 0.15 |
ENST00000589400.1
ENST00000590839.1 ENST00000592079.1 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chrX_+_78003204 | 0.15 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr20_-_50418947 | 0.15 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr15_+_63682335 | 0.15 |
ENST00000559379.1
ENST00000559821.1 |
RP11-321G12.1
|
RP11-321G12.1 |
| chr4_-_175204765 | 0.15 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr3_+_182983126 | 0.15 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr9_-_139965000 | 0.15 |
ENST00000409687.3
|
SAPCD2
|
suppressor APC domain containing 2 |
| chr8_-_42358742 | 0.15 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr16_-_29934558 | 0.14 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr3_+_186353756 | 0.14 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr4_+_129732419 | 0.14 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr1_-_242162375 | 0.14 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr17_+_43238438 | 0.14 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr8_+_97773202 | 0.14 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr7_-_83824449 | 0.13 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_7307358 | 0.13 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr16_+_14280742 | 0.13 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr18_-_33702078 | 0.13 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr15_-_53002007 | 0.13 |
ENST00000561490.1
|
FAM214A
|
family with sequence similarity 214, member A |
| chr11_+_72983246 | 0.13 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr7_+_93652116 | 0.13 |
ENST00000415536.1
|
AC003092.1
|
AC003092.1 |
| chr17_-_38928414 | 0.13 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr11_+_101918153 | 0.13 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr20_+_43538756 | 0.13 |
ENST00000537323.1
ENST00000217073.2 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr12_+_7014126 | 0.13 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr16_+_30675654 | 0.13 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chrX_+_100224676 | 0.13 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr1_+_154401791 | 0.13 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr11_+_47293795 | 0.13 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr17_+_67498538 | 0.12 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr17_-_46799872 | 0.12 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr4_+_183065793 | 0.12 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr1_-_89357179 | 0.12 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr15_+_76016293 | 0.12 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr3_+_107364769 | 0.12 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr16_+_69345243 | 0.12 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr3_+_111393501 | 0.12 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr12_+_48592134 | 0.12 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chrX_-_73512358 | 0.12 |
ENST00000602776.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chrX_+_54947229 | 0.12 |
ENST00000442098.1
ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO
|
trophinin |
| chr20_-_50418972 | 0.12 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr1_-_78444738 | 0.12 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr7_-_44122063 | 0.12 |
ENST00000335195.6
ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM
|
polymerase (DNA directed), mu |
| chr17_-_33446820 | 0.11 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr3_+_138340049 | 0.11 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr10_-_116418053 | 0.11 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr8_-_29120580 | 0.11 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr3_-_12587055 | 0.11 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr2_+_11682790 | 0.11 |
ENST00000389825.3
ENST00000381483.2 |
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr1_+_236958554 | 0.11 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr11_+_62496114 | 0.11 |
ENST00000532583.1
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr7_-_107883678 | 0.11 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chrX_-_73512411 | 0.11 |
ENST00000602576.1
ENST00000429124.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr17_-_63822563 | 0.11 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr4_+_159727272 | 0.11 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr16_+_24741013 | 0.11 |
ENST00000315183.7
ENST00000395799.3 |
TNRC6A
|
trinucleotide repeat containing 6A |
| chr4_+_119809984 | 0.11 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr11_-_93474645 | 0.11 |
ENST00000532455.1
|
TAF1D
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
| chr17_+_33914424 | 0.11 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr9_+_109685630 | 0.10 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr1_-_50889155 | 0.10 |
ENST00000404795.3
|
DMRTA2
|
DMRT-like family A2 |
| chr2_-_203735484 | 0.10 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr2_+_38152462 | 0.10 |
ENST00000354545.2
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr20_+_43538692 | 0.10 |
ENST00000217074.4
ENST00000255136.3 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr8_+_32579341 | 0.10 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr15_+_58702742 | 0.10 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr1_+_179923873 | 0.10 |
ENST00000367607.3
ENST00000491495.2 |
CEP350
|
centrosomal protein 350kDa |
| chr13_-_28545276 | 0.10 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr6_-_75994025 | 0.10 |
ENST00000518161.1
|
TMEM30A
|
transmembrane protein 30A |
| chr7_+_99717230 | 0.10 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr7_-_27169801 | 0.10 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr13_-_110438914 | 0.10 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr2_+_7073174 | 0.10 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr17_+_79495397 | 0.10 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr20_+_34020827 | 0.10 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr3_-_44519131 | 0.10 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr12_-_56694083 | 0.09 |
ENST00000552688.1
ENST00000548041.1 ENST00000551137.1 ENST00000551968.1 ENST00000542324.2 ENST00000546930.1 ENST00000549221.1 ENST00000550159.1 ENST00000550734.1 |
CS
|
citrate synthase |
| chr11_-_33913708 | 0.09 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chrX_-_77225135 | 0.09 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr12_+_107712173 | 0.09 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr12_-_10022735 | 0.09 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_-_118797475 | 0.09 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr14_-_37051798 | 0.09 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr17_+_48823975 | 0.09 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr1_+_202317815 | 0.09 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr2_-_17981462 | 0.09 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr17_-_18430160 | 0.09 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr3_-_33686925 | 0.09 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_-_118023594 | 0.09 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr2_+_135596180 | 0.09 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr7_+_6713376 | 0.09 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr3_-_141747950 | 0.09 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_28410128 | 0.09 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr12_+_106751436 | 0.09 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr10_+_7745303 | 0.09 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chrX_+_71401526 | 0.09 |
ENST00000218432.5
ENST00000423432.2 ENST00000373669.2 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr4_-_74486217 | 0.09 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_+_128502871 | 0.08 |
ENST00000249289.4
|
ATP6V1F
|
ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F |
| chr5_-_16916624 | 0.08 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr10_-_27529486 | 0.08 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr7_-_77427676 | 0.08 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr17_-_38821373 | 0.08 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr7_-_28220354 | 0.08 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr8_-_124553437 | 0.08 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr4_+_95917383 | 0.08 |
ENST00000512312.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chrX_+_10031499 | 0.08 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr9_-_107690420 | 0.08 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr6_-_127840336 | 0.08 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr12_-_118628350 | 0.08 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr4_-_8873531 | 0.08 |
ENST00000400677.3
|
HMX1
|
H6 family homeobox 1 |
| chr1_-_48866517 | 0.08 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr6_+_142623758 | 0.08 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr18_-_68004529 | 0.08 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr11_-_104827425 | 0.08 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr20_+_33292507 | 0.08 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr3_-_98619999 | 0.07 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr4_-_153601136 | 0.07 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr12_-_28123206 | 0.07 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr14_+_100485712 | 0.07 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr14_-_39639523 | 0.07 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr1_+_145439306 | 0.07 |
ENST00000425134.1
|
TXNIP
|
thioredoxin interacting protein |
| chr3_-_143567262 | 0.07 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr4_-_119759795 | 0.07 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr20_-_50419055 | 0.07 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr17_+_46970134 | 0.07 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr10_-_5046042 | 0.07 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr1_+_210501589 | 0.07 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr5_-_150473127 | 0.07 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr5_+_66300464 | 0.07 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_170440844 | 0.07 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr2_-_183387064 | 0.07 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_+_73455788 | 0.07 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chrX_+_10126488 | 0.07 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr16_+_53412368 | 0.07 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr4_-_25865159 | 0.07 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr10_+_13652047 | 0.07 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr10_+_91461413 | 0.07 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr5_-_13944652 | 0.07 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr15_-_37393406 | 0.07 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr13_-_30881134 | 0.07 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr17_-_74163159 | 0.07 |
ENST00000591615.1
|
RNF157
|
ring finger protein 157 |
| chr7_+_12726623 | 0.06 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr2_+_28618532 | 0.06 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr12_-_94673956 | 0.06 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr4_+_146539415 | 0.06 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr10_+_111765562 | 0.06 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr13_-_41593425 | 0.06 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr3_-_123339343 | 0.06 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.2 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.1 | 0.5 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.0 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.1 | 0.2 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.0 | GO:0031782 | melanocortin receptor binding(GO:0031779) corticotropin hormone receptor binding(GO:0031780) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |