Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
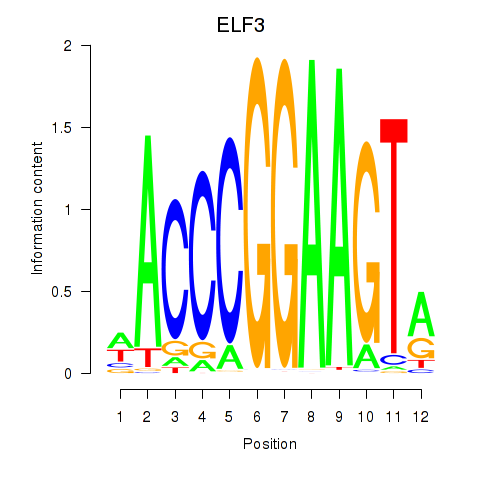
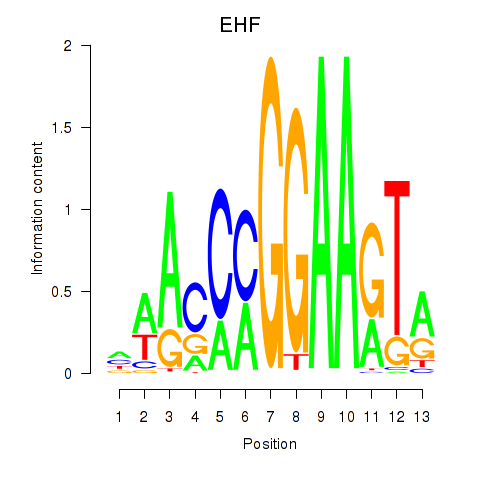
Results for ELF3_EHF
Z-value: 0.70
Transcription factors associated with ELF3_EHF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ELF3
|
ENSG00000163435.11 | E74 like ETS transcription factor 3 |
|
EHF
|
ENSG00000135373.8 | ETS homologous factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EHF | hg19_v2_chr11_+_34664014_34664174 | -0.97 | 3.3e-02 | Click! |
| ELF3 | hg19_v2_chr1_+_201979645_201979721 | 0.27 | 7.3e-01 | Click! |
Activity profile of ELF3_EHF motif
Sorted Z-values of ELF3_EHF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_37880543 | 0.36 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_-_25122785 | 0.36 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr1_-_762885 | 0.33 |
ENST00000536430.1
ENST00000473798.1 |
LINC00115
|
long intergenic non-protein coding RNA 115 |
| chrX_-_54069253 | 0.31 |
ENST00000425862.1
ENST00000433120.1 |
PHF8
|
PHD finger protein 8 |
| chr1_-_12679171 | 0.30 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr9_-_138391692 | 0.27 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr1_+_43637996 | 0.27 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr16_+_88636875 | 0.27 |
ENST00000569435.1
|
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr20_-_48532046 | 0.26 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr2_-_107502456 | 0.25 |
ENST00000419159.2
|
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
| chr3_-_52322019 | 0.23 |
ENST00000463624.1
|
WDR82
|
WD repeat domain 82 |
| chr6_-_109761707 | 0.23 |
ENST00000520723.1
ENST00000518648.1 ENST00000417394.2 |
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr15_-_55611306 | 0.23 |
ENST00000563262.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_+_175352114 | 0.23 |
ENST00000444196.1
ENST00000417038.1 ENST00000606406.1 |
AC010894.3
|
AC010894.3 |
| chr15_+_45879595 | 0.22 |
ENST00000565216.1
|
BLOC1S6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr2_+_65663812 | 0.22 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr12_-_12509929 | 0.22 |
ENST00000381800.2
|
LOH12CR2
|
loss of heterozygosity, 12, chromosomal region 2 (non-protein coding) |
| chr11_+_5710919 | 0.22 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr11_+_117198801 | 0.20 |
ENST00000527609.1
ENST00000533570.1 |
CEP164
|
centrosomal protein 164kDa |
| chr15_+_23810903 | 0.20 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr20_-_34287220 | 0.20 |
ENST00000306750.3
|
NFS1
|
NFS1 cysteine desulfurase |
| chr8_-_28347737 | 0.18 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr6_+_33257427 | 0.18 |
ENST00000463584.1
|
PFDN6
|
prefoldin subunit 6 |
| chr6_+_33257346 | 0.18 |
ENST00000374606.5
ENST00000374610.2 ENST00000374607.1 |
PFDN6
|
prefoldin subunit 6 |
| chr17_+_34958001 | 0.18 |
ENST00000250156.7
|
MRM1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr20_+_43104541 | 0.18 |
ENST00000372906.2
ENST00000456317.1 |
TTPAL
|
tocopherol (alpha) transfer protein-like |
| chr11_-_93276514 | 0.18 |
ENST00000526869.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr14_-_75083313 | 0.18 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr11_-_119993734 | 0.17 |
ENST00000533302.1
|
TRIM29
|
tripartite motif containing 29 |
| chr9_-_134615326 | 0.17 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr10_-_112255945 | 0.17 |
ENST00000609514.1
ENST00000607952.1 |
RP11-525A16.4
|
RP11-525A16.4 |
| chrX_+_56644819 | 0.17 |
ENST00000446028.1
|
RP11-431N15.2
|
RP11-431N15.2 |
| chr1_+_213224572 | 0.16 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr2_+_217363559 | 0.16 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr11_+_64008525 | 0.16 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr7_+_106415457 | 0.15 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr12_+_64845864 | 0.15 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr16_+_4845379 | 0.15 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr17_+_38376042 | 0.15 |
ENST00000583130.1
ENST00000584296.1 |
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr14_-_21852119 | 0.15 |
ENST00000555943.1
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae) |
| chr17_-_79269067 | 0.15 |
ENST00000288439.5
ENST00000374759.3 |
SLC38A10
|
solute carrier family 38, member 10 |
| chr3_-_139108463 | 0.14 |
ENST00000512242.1
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr2_+_58274001 | 0.14 |
ENST00000428021.1
|
VRK2
|
vaccinia related kinase 2 |
| chr3_-_42003613 | 0.14 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4 |
| chr2_+_66662249 | 0.14 |
ENST00000560281.2
|
MEIS1
|
Meis homeobox 1 |
| chr6_-_31620095 | 0.14 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr11_+_64008443 | 0.14 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr14_-_24701539 | 0.14 |
ENST00000534348.1
ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1
NEDD8
|
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr8_+_100025476 | 0.13 |
ENST00000355155.1
ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast) |
| chr1_-_154946792 | 0.13 |
ENST00000412170.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr12_-_80084333 | 0.13 |
ENST00000552637.1
|
PAWR
|
PRKC, apoptosis, WT1, regulator |
| chr1_+_169337412 | 0.13 |
ENST00000426663.1
|
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr3_-_171528227 | 0.13 |
ENST00000356327.5
ENST00000342215.6 ENST00000340989.4 ENST00000351298.4 |
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr19_-_19384055 | 0.13 |
ENST00000389363.4
|
TM6SF2
|
transmembrane 6 superfamily member 2 |
| chr19_-_53662257 | 0.12 |
ENST00000599096.1
ENST00000334197.7 ENST00000597183.1 ENST00000601804.1 ENST00000601469.2 ENST00000452676.2 |
ZNF347
|
zinc finger protein 347 |
| chr4_-_84255935 | 0.12 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr12_+_44152740 | 0.12 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr2_+_71295416 | 0.12 |
ENST00000455662.2
ENST00000531934.1 |
NAGK
|
N-acetylglucosamine kinase |
| chr15_+_55611401 | 0.12 |
ENST00000566999.1
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr3_-_57113281 | 0.12 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr11_-_69490135 | 0.12 |
ENST00000542341.1
|
ORAOV1
|
oral cancer overexpressed 1 |
| chr12_-_123215306 | 0.12 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr19_-_6393216 | 0.12 |
ENST00000595047.1
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa |
| chr19_-_54618650 | 0.12 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr15_-_45459704 | 0.12 |
ENST00000558039.1
|
CTD-2651B20.1
|
CTD-2651B20.1 |
| chr2_+_136499287 | 0.12 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr20_-_18774614 | 0.11 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr1_-_150978953 | 0.11 |
ENST00000493834.2
|
FAM63A
|
family with sequence similarity 63, member A |
| chr11_+_61891445 | 0.11 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr1_+_3541543 | 0.11 |
ENST00000378344.2
ENST00000344579.5 |
TPRG1L
|
tumor protein p63 regulated 1-like |
| chr11_-_9336117 | 0.11 |
ENST00000527813.1
ENST00000533723.1 |
TMEM41B
|
transmembrane protein 41B |
| chr18_+_74240756 | 0.11 |
ENST00000584910.1
ENST00000582452.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr22_-_39150947 | 0.11 |
ENST00000411587.2
ENST00000420859.1 ENST00000452294.1 ENST00000456894.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr16_+_31085714 | 0.11 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr3_+_52321827 | 0.11 |
ENST00000473032.1
ENST00000305690.8 ENST00000354773.4 ENST00000471180.1 ENST00000436784.2 |
GLYCTK
|
glycerate kinase |
| chr16_-_28223229 | 0.11 |
ENST00000566073.1
|
XPO6
|
exportin 6 |
| chr2_+_71295766 | 0.11 |
ENST00000533981.1
|
NAGK
|
N-acetylglucosamine kinase |
| chr17_-_27277615 | 0.11 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr2_+_162165038 | 0.10 |
ENST00000437630.1
|
PSMD14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr15_-_43663214 | 0.10 |
ENST00000561661.1
|
ZSCAN29
|
zinc finger and SCAN domain containing 29 |
| chr9_-_4679419 | 0.10 |
ENST00000609131.1
ENST00000607997.1 |
RP11-6J24.6
|
RP11-6J24.6 |
| chr12_-_118796971 | 0.10 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr14_-_55738788 | 0.10 |
ENST00000556183.1
|
RP11-665C16.6
|
RP11-665C16.6 |
| chr7_+_44240520 | 0.10 |
ENST00000496112.1
ENST00000223369.2 |
YKT6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr8_-_94753202 | 0.10 |
ENST00000521947.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr6_-_159420780 | 0.10 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr14_+_24701628 | 0.10 |
ENST00000355299.4
ENST00000559836.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr1_-_16556038 | 0.10 |
ENST00000375605.2
|
C1orf134
|
chromosome 1 open reading frame 134 |
| chr7_+_86781916 | 0.10 |
ENST00000579592.1
ENST00000434534.1 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr11_-_118889323 | 0.10 |
ENST00000527673.1
|
RPS25
|
ribosomal protein S25 |
| chr1_+_156308245 | 0.10 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr9_+_71736177 | 0.10 |
ENST00000606364.1
ENST00000453658.2 |
TJP2
|
tight junction protein 2 |
| chr5_+_140071011 | 0.10 |
ENST00000230771.3
ENST00000509299.1 ENST00000503873.1 ENST00000435019.2 ENST00000437649.2 ENST00000432671.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr5_-_93447333 | 0.10 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr15_-_65809625 | 0.10 |
ENST00000560436.1
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr1_-_150979333 | 0.10 |
ENST00000312210.5
|
FAM63A
|
family with sequence similarity 63, member A |
| chr11_+_313503 | 0.10 |
ENST00000528780.1
ENST00000328221.5 |
IFITM1
|
interferon induced transmembrane protein 1 |
| chr2_-_216003127 | 0.10 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr5_+_130506629 | 0.10 |
ENST00000510516.1
ENST00000507584.1 |
LYRM7
|
LYR motif containing 7 |
| chr5_-_137090028 | 0.09 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr15_-_74284613 | 0.09 |
ENST00000316911.6
ENST00000564777.1 ENST00000566081.1 ENST00000316900.5 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr19_-_53141584 | 0.09 |
ENST00000597161.1
ENST00000596930.1 ENST00000545872.1 ENST00000544146.1 ENST00000536937.1 ENST00000301096.3 |
ZNF83
|
zinc finger protein 83 |
| chr8_-_61429315 | 0.09 |
ENST00000530725.1
ENST00000532232.1 |
RP11-163N6.2
|
RP11-163N6.2 |
| chr6_-_111136299 | 0.09 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr1_-_222886526 | 0.09 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr14_-_54317532 | 0.09 |
ENST00000418927.1
|
AL162759.1
|
AL162759.1 |
| chr1_-_20126365 | 0.09 |
ENST00000294543.6
ENST00000375122.2 |
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr19_-_51014345 | 0.09 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr12_+_31227192 | 0.09 |
ENST00000535317.1
|
DDX11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr2_-_24149918 | 0.09 |
ENST00000439915.1
|
ATAD2B
|
ATPase family, AAA domain containing 2B |
| chr3_+_28390637 | 0.09 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chrX_-_67653291 | 0.09 |
ENST00000540071.1
|
OPHN1
|
oligophrenin 1 |
| chr19_-_54106751 | 0.09 |
ENST00000600193.1
|
CTB-167G5.5
|
Uncharacterized protein |
| chr8_+_105602961 | 0.09 |
ENST00000521923.1
|
RP11-127H5.1
|
Uncharacterized protein |
| chr16_+_2587998 | 0.09 |
ENST00000441549.3
ENST00000268673.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr14_-_105531759 | 0.09 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr10_-_375422 | 0.09 |
ENST00000434695.2
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr16_+_718147 | 0.09 |
ENST00000561929.1
|
RHOT2
|
ras homolog family member T2 |
| chr16_+_2588012 | 0.09 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr14_+_24701819 | 0.08 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr17_-_76837499 | 0.08 |
ENST00000592275.1
|
USP36
|
ubiquitin specific peptidase 36 |
| chr19_-_12662314 | 0.08 |
ENST00000339282.7
ENST00000596193.1 |
ZNF564
|
zinc finger protein 564 |
| chr1_+_154975110 | 0.08 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr21_-_46221684 | 0.08 |
ENST00000330942.5
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr2_+_201981663 | 0.08 |
ENST00000433445.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_201981527 | 0.08 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr3_+_57541975 | 0.08 |
ENST00000487257.1
ENST00000311180.8 |
PDE12
|
phosphodiesterase 12 |
| chr14_-_75593708 | 0.08 |
ENST00000557673.1
ENST00000238616.5 |
NEK9
|
NIMA-related kinase 9 |
| chr12_-_133405409 | 0.08 |
ENST00000545875.1
ENST00000456883.2 |
GOLGA3
|
golgin A3 |
| chr22_+_22020353 | 0.08 |
ENST00000456792.2
|
PPIL2
|
peptidylprolyl isomerase (cyclophilin)-like 2 |
| chr22_-_38245304 | 0.08 |
ENST00000609454.1
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr7_+_86781778 | 0.08 |
ENST00000432937.2
|
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr1_+_179335101 | 0.08 |
ENST00000508285.1
ENST00000511889.1 |
AXDND1
|
axonemal dynein light chain domain containing 1 |
| chr4_+_492985 | 0.08 |
ENST00000296306.7
ENST00000536264.1 ENST00000310340.5 ENST00000453061.2 ENST00000504346.1 ENST00000503111.1 ENST00000383028.4 ENST00000509768.1 |
PIGG
|
phosphatidylinositol glycan anchor biosynthesis, class G |
| chr1_-_197744763 | 0.08 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr22_-_38349552 | 0.08 |
ENST00000422191.1
ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23
|
chromosome 22 open reading frame 23 |
| chr14_-_74417096 | 0.08 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr11_-_58345569 | 0.08 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr2_+_38152462 | 0.08 |
ENST00000354545.2
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr15_-_52861157 | 0.08 |
ENST00000564163.1
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr2_-_85555086 | 0.08 |
ENST00000444342.2
ENST00000409232.3 ENST00000409015.1 |
TGOLN2
|
trans-golgi network protein 2 |
| chr17_-_55162360 | 0.08 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr21_-_33975547 | 0.08 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr17_-_11900689 | 0.08 |
ENST00000322748.3
ENST00000454073.3 ENST00000580903.1 ENST00000580306.2 |
ZNF18
|
zinc finger protein 18 |
| chr18_+_11851383 | 0.08 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr22_+_38004942 | 0.08 |
ENST00000439161.1
ENST00000449944.1 ENST00000411501.1 ENST00000453208.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr5_+_49962495 | 0.07 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_-_238499725 | 0.07 |
ENST00000264601.3
|
RAB17
|
RAB17, member RAS oncogene family |
| chr1_+_44889697 | 0.07 |
ENST00000443020.2
|
RNF220
|
ring finger protein 220 |
| chr11_-_64885111 | 0.07 |
ENST00000528598.1
ENST00000310597.4 |
ZNHIT2
|
zinc finger, HIT-type containing 2 |
| chr14_+_24702073 | 0.07 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr16_-_19729453 | 0.07 |
ENST00000564480.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr3_-_42003479 | 0.07 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr8_-_27168737 | 0.07 |
ENST00000521253.1
ENST00000305364.4 |
TRIM35
|
tripartite motif containing 35 |
| chr5_+_40841410 | 0.07 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr21_+_46359907 | 0.07 |
ENST00000291634.6
ENST00000397826.3 ENST00000458015.1 |
FAM207A
|
family with sequence similarity 207, member A |
| chr1_+_178994939 | 0.07 |
ENST00000440702.1
|
FAM20B
|
family with sequence similarity 20, member B |
| chr11_-_62389449 | 0.07 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr16_-_11680791 | 0.07 |
ENST00000571976.1
ENST00000413364.2 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr6_-_99873145 | 0.07 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr22_+_27068704 | 0.07 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr1_+_213123862 | 0.07 |
ENST00000366966.2
ENST00000366964.3 |
VASH2
|
vasohibin 2 |
| chr19_-_12708113 | 0.07 |
ENST00000440366.1
|
ZNF490
|
zinc finger protein 490 |
| chr15_-_65809991 | 0.07 |
ENST00000559526.1
ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8
|
dipeptidyl-peptidase 8 |
| chr19_-_18654828 | 0.07 |
ENST00000609656.1
ENST00000597611.3 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr4_+_166128836 | 0.07 |
ENST00000511305.1
|
KLHL2
|
kelch-like family member 2 |
| chr7_-_944631 | 0.07 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr2_-_75788038 | 0.07 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr14_-_71107921 | 0.07 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr16_+_20817839 | 0.07 |
ENST00000348433.6
ENST00000568501.1 ENST00000566276.1 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr11_+_70244510 | 0.07 |
ENST00000346329.3
ENST00000301843.8 ENST00000376561.3 |
CTTN
|
cortactin |
| chr19_+_35739782 | 0.07 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr1_-_197744390 | 0.07 |
ENST00000367396.3
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr2_+_30670209 | 0.07 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr5_+_140071178 | 0.07 |
ENST00000508522.1
ENST00000448069.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr12_+_82752647 | 0.07 |
ENST00000550058.1
|
METTL25
|
methyltransferase like 25 |
| chr6_+_36853607 | 0.07 |
ENST00000480824.2
ENST00000355190.3 ENST00000373685.1 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr17_+_38375528 | 0.07 |
ENST00000583268.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr19_-_53400813 | 0.07 |
ENST00000595635.1
ENST00000594741.1 ENST00000597111.1 ENST00000593618.1 ENST00000597909.1 |
ZNF320
|
zinc finger protein 320 |
| chr6_-_41747595 | 0.07 |
ENST00000373018.3
|
FRS3
|
fibroblast growth factor receptor substrate 3 |
| chr11_-_69490073 | 0.07 |
ENST00000535657.1
ENST00000539414.1 ENST00000536870.1 ENST00000538554.2 ENST00000279147.4 |
ORAOV1
|
oral cancer overexpressed 1 |
| chr11_-_62389577 | 0.07 |
ENST00000534715.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr1_+_43855545 | 0.07 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr16_+_2587965 | 0.07 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr2_+_71295733 | 0.07 |
ENST00000443938.2
ENST00000244204.6 |
NAGK
|
N-acetylglucosamine kinase |
| chr1_-_228353112 | 0.07 |
ENST00000366713.1
|
IBA57-AS1
|
IBA57 antisense RNA 1 (head to head) |
| chr1_+_100598742 | 0.07 |
ENST00000370139.1
|
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr12_-_94673956 | 0.07 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr8_+_27168988 | 0.07 |
ENST00000397501.1
ENST00000338238.4 ENST00000544172.1 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr1_+_76251912 | 0.07 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr1_+_24285599 | 0.07 |
ENST00000471915.1
|
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr12_-_133405288 | 0.07 |
ENST00000204726.3
|
GOLGA3
|
golgin A3 |
| chr7_-_38370536 | 0.07 |
ENST00000390343.2
|
TRGV8
|
T cell receptor gamma variable 8 |
| chr17_-_43568062 | 0.07 |
ENST00000421073.2
ENST00000584420.1 ENST00000589780.1 ENST00000430334.3 |
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr5_+_180650271 | 0.07 |
ENST00000351937.5
ENST00000315073.5 |
TRIM41
|
tripartite motif containing 41 |
| chr12_+_76653682 | 0.07 |
ENST00000553247.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr7_-_140178726 | 0.06 |
ENST00000480552.1
|
MKRN1
|
makorin ring finger protein 1 |
| chr10_-_22292675 | 0.06 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr14_+_20187174 | 0.06 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr12_+_50794947 | 0.06 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ELF3_EHF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.2 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 0.1 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.1 | 0.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.2 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.3 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.0 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.1 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.0 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.0 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.0 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.0 | GO:0002415 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.0 | GO:0097198 | cell migration involved in vasculogenesis(GO:0035441) histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.0 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.0 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0009217 | purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 0.0 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.0 | GO:0031592 | centrosomal corona(GO:0031592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.2 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.1 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0016731 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |