Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
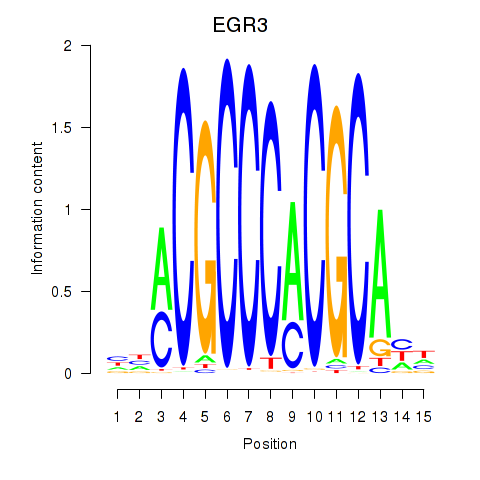
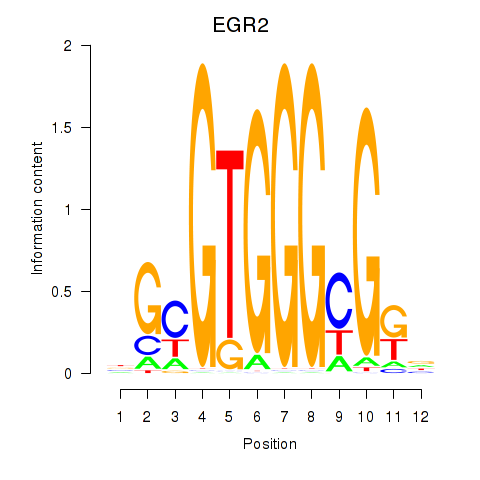
Results for EGR3_EGR2
Z-value: 0.78
Transcription factors associated with EGR3_EGR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR3
|
ENSG00000179388.8 | early growth response 3 |
|
EGR2
|
ENSG00000122877.9 | early growth response 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR2 | hg19_v2_chr10_-_64576105_64576133 | -0.96 | 4.2e-02 | Click! |
| EGR3 | hg19_v2_chr8_-_22550815_22550844 | -0.77 | 2.3e-01 | Click! |
Activity profile of EGR3_EGR2 motif
Sorted Z-values of EGR3_EGR2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_76649753 | 0.77 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr6_+_12012170 | 0.51 |
ENST00000487103.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr19_-_49622348 | 0.44 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr2_-_98612379 | 0.43 |
ENST00000425805.2
|
TMEM131
|
transmembrane protein 131 |
| chr2_+_149402009 | 0.37 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr19_+_46144884 | 0.35 |
ENST00000593161.1
|
AC006132.1
|
chromosome 19 open reading frame 83 |
| chr1_-_85156417 | 0.34 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr11_+_64009072 | 0.32 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr4_-_83295296 | 0.32 |
ENST00000507010.1
ENST00000503822.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr16_-_31021717 | 0.32 |
ENST00000565419.1
|
STX1B
|
syntaxin 1B |
| chr19_-_49371711 | 0.31 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr15_-_72410455 | 0.31 |
ENST00000569314.1
|
MYO9A
|
myosin IXA |
| chrX_-_48814810 | 0.31 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr11_+_117198801 | 0.30 |
ENST00000527609.1
ENST00000533570.1 |
CEP164
|
centrosomal protein 164kDa |
| chr19_-_46145696 | 0.29 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr8_-_101322132 | 0.29 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr14_-_51027838 | 0.27 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr16_-_54962704 | 0.27 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr7_+_5465382 | 0.26 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr9_+_127020202 | 0.26 |
ENST00000373600.3
ENST00000320246.5 |
NEK6
|
NIMA-related kinase 6 |
| chr2_+_191513789 | 0.26 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr15_-_71146460 | 0.25 |
ENST00000344870.4
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr2_+_220306745 | 0.24 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr19_+_1261106 | 0.24 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr4_-_83351294 | 0.23 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr12_-_57634475 | 0.23 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr2_+_177502438 | 0.22 |
ENST00000443670.1
|
AC017048.4
|
long intergenic non-protein coding RNA 1117 |
| chr5_-_176924562 | 0.22 |
ENST00000359895.2
ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr17_+_46985823 | 0.21 |
ENST00000508468.2
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr15_+_76030311 | 0.20 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr2_-_74648702 | 0.20 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr4_-_76649546 | 0.20 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr12_+_77158021 | 0.19 |
ENST00000550876.1
|
ZDHHC17
|
zinc finger, DHHC-type containing 17 |
| chr2_-_239149300 | 0.19 |
ENST00000436051.1
|
HES6
|
hes family bHLH transcription factor 6 |
| chr6_-_110500826 | 0.19 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr11_-_72853267 | 0.19 |
ENST00000409418.4
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr4_-_83295103 | 0.18 |
ENST00000313899.7
ENST00000352301.4 ENST00000509107.1 ENST00000353341.4 ENST00000541060.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr9_-_14322319 | 0.18 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr11_+_73498973 | 0.18 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr5_-_178054014 | 0.18 |
ENST00000520957.1
|
CLK4
|
CDC-like kinase 4 |
| chr6_-_33266492 | 0.18 |
ENST00000425946.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr17_-_79008373 | 0.18 |
ENST00000577066.1
ENST00000573167.1 |
BAIAP2-AS1
|
BAIAP2 antisense RNA 1 (head to head) |
| chr1_+_23695680 | 0.17 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr15_+_40733387 | 0.17 |
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr19_+_49617581 | 0.17 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr19_+_10400615 | 0.17 |
ENST00000221980.4
|
ICAM5
|
intercellular adhesion molecule 5, telencephalin |
| chr2_+_187350973 | 0.17 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr4_-_78740769 | 0.17 |
ENST00000512485.1
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr11_-_65640071 | 0.16 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr7_+_151038850 | 0.16 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr8_+_145490549 | 0.16 |
ENST00000340695.2
|
SCXA
|
scleraxis homolog A (mouse) |
| chr15_-_40574787 | 0.16 |
ENST00000434396.1
|
ANKRD63
|
ankyrin repeat domain 63 |
| chr3_-_149688971 | 0.16 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr1_-_16302608 | 0.16 |
ENST00000375743.4
ENST00000375733.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chrX_-_48814278 | 0.15 |
ENST00000455452.1
|
OTUD5
|
OTU domain containing 5 |
| chr12_-_6715808 | 0.15 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr3_+_50192833 | 0.14 |
ENST00000426511.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr2_+_191513959 | 0.14 |
ENST00000337386.5
ENST00000357215.5 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr4_-_36246060 | 0.14 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr3_-_171177852 | 0.14 |
ENST00000284483.8
ENST00000475336.1 ENST00000357327.5 ENST00000460047.1 ENST00000488470.1 ENST00000470834.1 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr19_-_14201776 | 0.14 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr10_-_27444143 | 0.14 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr17_-_4710288 | 0.14 |
ENST00000571067.1
|
RP11-81A22.5
|
RP11-81A22.5 |
| chr1_-_197744763 | 0.14 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr16_-_28608364 | 0.14 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr10_-_95462265 | 0.13 |
ENST00000536233.1
ENST00000359204.4 ENST00000371430.2 ENST00000394100.2 |
FRA10AC1
|
fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 |
| chr7_+_151038785 | 0.13 |
ENST00000413040.2
ENST00000568733.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr3_+_184056614 | 0.13 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr7_+_70229899 | 0.13 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr20_+_35201993 | 0.13 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr2_+_219724544 | 0.13 |
ENST00000233948.3
|
WNT6
|
wingless-type MMTV integration site family, member 6 |
| chr9_+_35829208 | 0.13 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr8_-_57123815 | 0.13 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr6_-_31633624 | 0.13 |
ENST00000375895.2
ENST00000375900.4 |
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr10_-_127408011 | 0.12 |
ENST00000531977.1
ENST00000527483.1 ENST00000525909.1 ENST00000528844.1 ENST00000423178.2 |
RP11-383C5.4
|
RP11-383C5.4 |
| chrX_+_155227371 | 0.12 |
ENST00000369423.2
ENST00000540897.1 |
IL9R
|
interleukin 9 receptor |
| chr19_-_663147 | 0.12 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chrX_-_19905577 | 0.12 |
ENST00000379697.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr4_+_26859300 | 0.12 |
ENST00000494628.2
|
STIM2
|
stromal interaction molecule 2 |
| chr1_+_145726886 | 0.12 |
ENST00000443667.1
|
PDZK1
|
PDZ domain containing 1 |
| chrX_+_155227246 | 0.12 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr10_-_18948208 | 0.12 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr16_-_850723 | 0.12 |
ENST00000248150.4
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13 |
| chr11_+_114310164 | 0.12 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr14_+_21566980 | 0.12 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr20_+_2673383 | 0.12 |
ENST00000380648.4
ENST00000342725.5 |
EBF4
|
early B-cell factor 4 |
| chr12_+_69864129 | 0.12 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr17_-_73178599 | 0.12 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr17_+_36858694 | 0.12 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr3_-_88108212 | 0.11 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr3_-_135915146 | 0.11 |
ENST00000473093.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr6_-_42110342 | 0.11 |
ENST00000356542.5
|
C6orf132
|
chromosome 6 open reading frame 132 |
| chr12_+_70760056 | 0.11 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr6_+_134274354 | 0.11 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr3_-_113415441 | 0.11 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr5_-_142784101 | 0.11 |
ENST00000503201.1
ENST00000502892.1 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_+_91526668 | 0.11 |
ENST00000521334.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr4_+_38869298 | 0.11 |
ENST00000510213.1
ENST00000515037.1 |
FAM114A1
|
family with sequence similarity 114, member A1 |
| chr2_+_105953972 | 0.11 |
ENST00000410049.1
|
C2orf49
|
chromosome 2 open reading frame 49 |
| chr3_-_171178157 | 0.11 |
ENST00000465393.1
ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr12_-_105352047 | 0.11 |
ENST00000432951.1
ENST00000415674.1 ENST00000424946.1 |
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr9_-_15510287 | 0.11 |
ENST00000397519.2
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr19_+_11457232 | 0.11 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr17_-_4890919 | 0.11 |
ENST00000572543.1
ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2
|
calmodulin binding transcription activator 2 |
| chrX_-_47509994 | 0.11 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chr17_-_62502399 | 0.11 |
ENST00000450599.2
ENST00000585060.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr9_-_101017900 | 0.10 |
ENST00000375066.5
|
TBC1D2
|
TBC1 domain family, member 2 |
| chr11_+_8704748 | 0.10 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr15_-_34502197 | 0.10 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr11_-_110167331 | 0.10 |
ENST00000534683.1
|
RDX
|
radixin |
| chr2_+_233562015 | 0.10 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr1_-_23670752 | 0.10 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr6_-_110501126 | 0.10 |
ENST00000368938.1
|
WASF1
|
WAS protein family, member 1 |
| chr6_-_32157947 | 0.10 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr15_-_45480153 | 0.10 |
ENST00000560471.1
ENST00000560540.1 |
SHF
|
Src homology 2 domain containing F |
| chrX_-_49056635 | 0.10 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr2_+_111878483 | 0.10 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr15_-_89089860 | 0.10 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr6_-_31088214 | 0.10 |
ENST00000376288.2
|
CDSN
|
corneodesmosin |
| chr2_-_48132814 | 0.10 |
ENST00000316377.4
ENST00000378314.3 |
FBXO11
|
F-box protein 11 |
| chr15_-_42783303 | 0.10 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr13_-_52026730 | 0.10 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr19_-_38878632 | 0.10 |
ENST00000586599.1
ENST00000334928.6 ENST00000587676.1 |
GGN
|
gametogenetin |
| chr10_+_91461413 | 0.09 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr7_+_90032821 | 0.09 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr7_-_43909090 | 0.09 |
ENST00000317534.5
|
MRPS24
|
mitochondrial ribosomal protein S24 |
| chrX_-_128788914 | 0.09 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr10_+_114709999 | 0.09 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr20_-_4229721 | 0.09 |
ENST00000379453.4
|
ADRA1D
|
adrenoceptor alpha 1D |
| chr9_+_96338860 | 0.09 |
ENST00000375376.4
|
PHF2
|
PHD finger protein 2 |
| chr16_-_67969888 | 0.09 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr15_+_52311398 | 0.09 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr6_+_2245982 | 0.09 |
ENST00000530346.1
ENST00000524770.1 ENST00000532124.1 ENST00000531092.1 ENST00000456943.2 ENST00000529893.1 |
GMDS-AS1
|
GMDS antisense RNA 1 (head to head) |
| chr17_+_20059358 | 0.09 |
ENST00000536879.1
ENST00000395522.2 ENST00000395525.3 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr2_-_222436988 | 0.09 |
ENST00000409854.1
ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4
|
EPH receptor A4 |
| chr9_+_108210279 | 0.09 |
ENST00000374716.4
ENST00000374710.3 ENST00000481272.1 ENST00000484973.1 ENST00000394926.3 ENST00000539376.1 |
FSD1L
|
fibronectin type III and SPRY domain containing 1-like |
| chr16_+_2286726 | 0.09 |
ENST00000382437.4
ENST00000569184.1 |
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr17_-_4890649 | 0.09 |
ENST00000361571.5
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr11_+_8704298 | 0.09 |
ENST00000531978.1
ENST00000524496.1 ENST00000532359.1 ENST00000530022.1 |
RPL27A
|
ribosomal protein L27a |
| chr9_+_127020503 | 0.09 |
ENST00000545174.1
ENST00000444973.1 ENST00000454453.1 |
NEK6
|
NIMA-related kinase 6 |
| chr1_+_178994939 | 0.09 |
ENST00000440702.1
|
FAM20B
|
family with sequence similarity 20, member B |
| chr22_+_36784632 | 0.09 |
ENST00000424761.1
|
RP4-633O19__A.1
|
RP4-633O19__A.1 |
| chr8_+_26149274 | 0.09 |
ENST00000522535.1
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr6_-_37665751 | 0.09 |
ENST00000297153.7
ENST00000434837.3 |
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr3_-_52002194 | 0.09 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr13_-_52027134 | 0.09 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr9_-_132597529 | 0.08 |
ENST00000372447.3
|
C9orf78
|
chromosome 9 open reading frame 78 |
| chr19_+_44037546 | 0.08 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr7_-_100808843 | 0.08 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr17_+_44668387 | 0.08 |
ENST00000576040.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chrX_+_117480036 | 0.08 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr10_+_22614547 | 0.08 |
ENST00000416820.1
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr5_-_133706695 | 0.08 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr15_-_72410109 | 0.08 |
ENST00000564571.1
|
MYO9A
|
myosin IXA |
| chr8_-_27469383 | 0.08 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr14_+_66974845 | 0.08 |
ENST00000459628.1
|
GPHN
|
gephyrin |
| chr15_-_83316711 | 0.08 |
ENST00000568128.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr14_-_75079294 | 0.08 |
ENST00000556359.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr16_-_54320675 | 0.08 |
ENST00000329734.3
|
IRX3
|
iroquois homeobox 3 |
| chr17_+_57697246 | 0.08 |
ENST00000580081.1
ENST00000579456.1 |
CLTC
|
clathrin, heavy chain (Hc) |
| chr7_-_27213893 | 0.08 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr11_+_114310237 | 0.08 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr1_-_67896009 | 0.08 |
ENST00000370990.5
|
SERBP1
|
SERPINE1 mRNA binding protein 1 |
| chr8_+_109455830 | 0.08 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr19_+_44037334 | 0.08 |
ENST00000314228.5
|
ZNF575
|
zinc finger protein 575 |
| chr14_+_70234642 | 0.08 |
ENST00000555349.1
ENST00000554021.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr13_-_45915221 | 0.08 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr19_-_33793430 | 0.08 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr19_-_37096139 | 0.08 |
ENST00000585983.1
ENST00000585960.1 ENST00000586115.1 |
ZNF529
|
zinc finger protein 529 |
| chr12_+_75728419 | 0.08 |
ENST00000378695.4
ENST00000312442.2 |
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr19_-_15311713 | 0.08 |
ENST00000601011.1
ENST00000263388.2 |
NOTCH3
|
notch 3 |
| chr5_-_176923803 | 0.08 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr6_-_166401442 | 0.08 |
ENST00000584179.1
|
LINC00473
|
long intergenic non-protein coding RNA 473 |
| chr6_-_46620522 | 0.08 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr11_+_61891445 | 0.08 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr17_-_62502639 | 0.08 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr7_+_43803790 | 0.08 |
ENST00000424330.1
|
BLVRA
|
biliverdin reductase A |
| chr10_-_32217717 | 0.08 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr13_+_111365602 | 0.08 |
ENST00000333219.7
|
ING1
|
inhibitor of growth family, member 1 |
| chr1_-_180991978 | 0.08 |
ENST00000542060.1
ENST00000258301.5 |
STX6
|
syntaxin 6 |
| chr19_+_18794470 | 0.08 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr16_-_28621298 | 0.08 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr11_-_75921780 | 0.08 |
ENST00000529461.1
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr5_+_135468516 | 0.07 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr11_+_101918153 | 0.07 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr16_+_3070356 | 0.07 |
ENST00000341627.5
ENST00000575124.1 ENST00000575836.1 |
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr8_-_57233103 | 0.07 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr21_+_35014706 | 0.07 |
ENST00000399353.1
ENST00000444491.1 ENST00000381318.3 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr19_-_51568324 | 0.07 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr4_+_76649797 | 0.07 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr11_-_65363259 | 0.07 |
ENST00000342202.4
|
KCNK7
|
potassium channel, subfamily K, member 7 |
| chr5_+_67584174 | 0.07 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr7_-_100808394 | 0.07 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr14_-_69658127 | 0.07 |
ENST00000556182.1
|
RP11-363J20.1
|
RP11-363J20.1 |
| chr5_-_16936340 | 0.07 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr19_+_55795493 | 0.07 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr19_-_10530784 | 0.07 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr6_+_133562472 | 0.07 |
ENST00000430974.2
ENST00000367895.5 ENST00000355167.3 ENST00000355286.6 |
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr9_+_116172958 | 0.07 |
ENST00000374165.1
|
C9orf43
|
chromosome 9 open reading frame 43 |
| chr15_-_43785274 | 0.07 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR3_EGR2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.1 | 0.3 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:1902230 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.1 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.2 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.0 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0072302 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.0 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:2001112 | negative regulation of hepatocyte growth factor receptor signaling pathway(GO:1902203) regulation of cellular response to hepatocyte growth factor stimulus(GO:2001112) negative regulation of cellular response to hepatocyte growth factor stimulus(GO:2001113) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.2 | GO:1901166 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.0 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.0 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.0 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.0 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.0 | GO:2000543 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.0 | GO:0015993 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.0 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.0 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.0 | GO:0097198 | cell migration involved in vasculogenesis(GO:0035441) histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.0 | GO:1902232 | regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.0 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.0 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.0 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.0 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.0 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.0 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |