Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
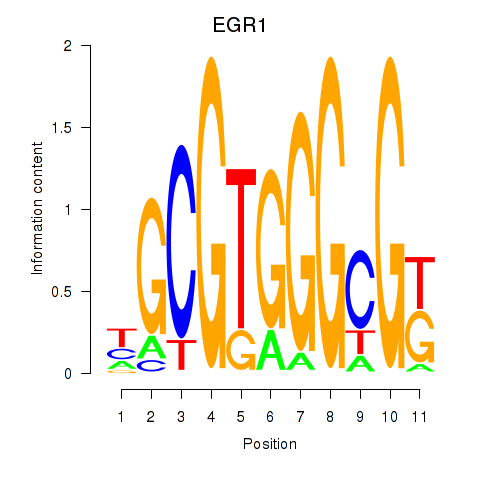
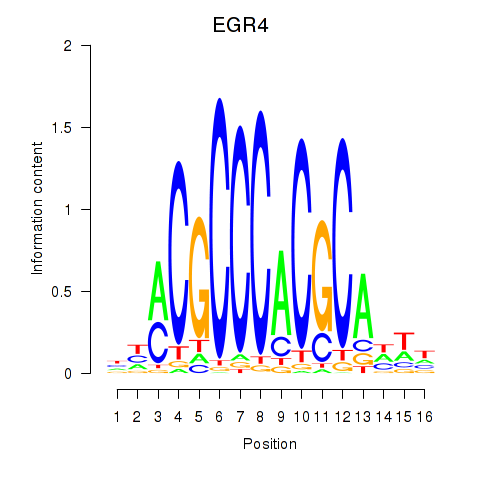
Results for EGR1_EGR4
Z-value: 0.48
Transcription factors associated with EGR1_EGR4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR1
|
ENSG00000120738.7 | early growth response 1 |
|
EGR4
|
ENSG00000135625.6 | early growth response 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR1 | hg19_v2_chr5_+_137801160_137801179 | 0.23 | 7.7e-01 | Click! |
Activity profile of EGR1_EGR4 motif
Sorted Z-values of EGR1_EGR4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_61108449 | 0.32 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr2_+_191513587 | 0.26 |
ENST00000416973.1
ENST00000426601.1 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr19_+_47778119 | 0.25 |
ENST00000552360.2
|
PRR24
|
proline rich 24 |
| chr17_-_77179487 | 0.25 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr1_-_9189144 | 0.22 |
ENST00000414642.2
|
GPR157
|
G protein-coupled receptor 157 |
| chr2_+_220306238 | 0.22 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr11_+_124609742 | 0.20 |
ENST00000284292.6
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr16_+_3070313 | 0.19 |
ENST00000326577.4
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr11_+_66624527 | 0.19 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr5_-_172662197 | 0.18 |
ENST00000521848.1
|
NKX2-5
|
NK2 homeobox 5 |
| chr17_+_72428266 | 0.18 |
ENST00000582473.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_+_124609823 | 0.17 |
ENST00000412681.2
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr1_+_38273988 | 0.16 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr1_+_38273818 | 0.15 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr9_-_34637806 | 0.15 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr5_+_421030 | 0.14 |
ENST00000506456.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr5_-_172662230 | 0.14 |
ENST00000424406.2
|
NKX2-5
|
NK2 homeobox 5 |
| chr19_-_46405861 | 0.14 |
ENST00000322217.5
|
MYPOP
|
Myb-related transcription factor, partner of profilin |
| chr16_+_2564254 | 0.14 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr2_+_242498135 | 0.14 |
ENST00000318407.3
|
BOK
|
BCL2-related ovarian killer |
| chr22_+_46546406 | 0.14 |
ENST00000440343.1
ENST00000415785.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr7_+_100860949 | 0.13 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr6_+_138188351 | 0.13 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr14_-_21566731 | 0.13 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr22_+_50354104 | 0.13 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr1_-_33647267 | 0.13 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr5_+_175223715 | 0.13 |
ENST00000515502.1
|
CPLX2
|
complexin 2 |
| chr5_+_133562095 | 0.12 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr19_-_54693401 | 0.12 |
ENST00000338624.6
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr5_+_172484377 | 0.12 |
ENST00000523161.1
|
CREBRF
|
CREB3 regulatory factor |
| chr15_+_74833518 | 0.11 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr17_-_7297833 | 0.11 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr11_-_64512803 | 0.11 |
ENST00000377489.1
ENST00000354024.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr2_-_27718052 | 0.11 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr19_+_589893 | 0.11 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr22_-_30722866 | 0.11 |
ENST00000403477.3
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr14_+_102027688 | 0.10 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr19_+_36132631 | 0.10 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr9_+_34652164 | 0.10 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr1_+_156698743 | 0.10 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr19_-_14201507 | 0.10 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr11_-_506739 | 0.10 |
ENST00000529306.1
ENST00000438658.2 ENST00000527485.1 ENST00000397615.2 ENST00000397614.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr17_-_42402138 | 0.10 |
ENST00000592857.1
ENST00000586016.1 ENST00000590194.1 ENST00000377095.5 ENST00000588049.1 ENST00000586633.1 ENST00000537904.2 ENST00000585636.1 ENST00000585523.1 ENST00000225308.8 |
SLC25A39
|
solute carrier family 25, member 39 |
| chr20_+_49348081 | 0.10 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr1_-_94312706 | 0.10 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_+_155051305 | 0.10 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr20_-_31172598 | 0.10 |
ENST00000201961.2
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr4_+_103422499 | 0.10 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr16_+_3014269 | 0.10 |
ENST00000575885.1
ENST00000571007.1 ENST00000319500.6 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr19_+_49617609 | 0.10 |
ENST00000221459.2
ENST00000486217.2 |
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr10_+_49514698 | 0.09 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr1_+_161494036 | 0.09 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chr6_+_11537910 | 0.09 |
ENST00000543875.1
|
TMEM170B
|
transmembrane protein 170B |
| chr4_-_90758227 | 0.09 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_+_54671047 | 0.09 |
ENST00000332822.4
|
NOG
|
noggin |
| chr8_+_145064233 | 0.09 |
ENST00000529301.1
ENST00000395068.4 |
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr18_+_3449821 | 0.09 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr19_-_54693521 | 0.09 |
ENST00000391754.1
ENST00000245615.1 ENST00000431666.2 |
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chrX_+_71996972 | 0.09 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chrX_-_47479246 | 0.09 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr20_+_35202909 | 0.09 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr2_-_128145498 | 0.09 |
ENST00000409179.2
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr19_-_1863567 | 0.09 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr14_-_20922960 | 0.09 |
ENST00000553640.1
ENST00000488532.2 |
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr16_+_30194916 | 0.09 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chr19_-_663277 | 0.09 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
| chrX_-_23926004 | 0.08 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr9_-_140196703 | 0.08 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chrX_+_117957741 | 0.08 |
ENST00000310164.2
|
ZCCHC12
|
zinc finger, CCHC domain containing 12 |
| chr1_+_65886326 | 0.08 |
ENST00000371059.3
ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR
|
leptin receptor |
| chr5_-_101632153 | 0.08 |
ENST00000310954.6
|
SLCO4C1
|
solute carrier organic anion transporter family, member 4C1 |
| chr19_+_10531150 | 0.08 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr14_-_23822080 | 0.08 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr11_-_507184 | 0.08 |
ENST00000533410.1
ENST00000354420.2 ENST00000397604.3 ENST00000531149.1 ENST00000356187.5 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr10_+_6625605 | 0.08 |
ENST00000414894.1
ENST00000449648.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr4_-_82393009 | 0.08 |
ENST00000436139.2
|
RASGEF1B
|
RasGEF domain family, member 1B |
| chr19_-_54693146 | 0.08 |
ENST00000414665.1
ENST00000453320.1 |
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr16_-_2264779 | 0.08 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr3_-_135916073 | 0.08 |
ENST00000481989.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr19_-_5567842 | 0.08 |
ENST00000587632.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr17_+_77751931 | 0.08 |
ENST00000310942.4
ENST00000269399.5 |
CBX2
|
chromobox homolog 2 |
| chr19_+_50879705 | 0.08 |
ENST00000598168.1
ENST00000411902.2 ENST00000253727.5 ENST00000597790.1 ENST00000597130.1 ENST00000599105.1 |
NR1H2
|
nuclear receptor subfamily 1, group H, member 2 |
| chr17_+_46185111 | 0.07 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr11_+_46403303 | 0.07 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr22_-_30722912 | 0.07 |
ENST00000215790.7
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr20_+_48429356 | 0.07 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr19_-_36523529 | 0.07 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr6_+_144471643 | 0.07 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr12_-_56223363 | 0.07 |
ENST00000546957.1
|
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr17_+_72772621 | 0.07 |
ENST00000335464.5
ENST00000417024.2 ENST00000578764.1 ENST00000582773.1 ENST00000582330.1 |
TMEM104
|
transmembrane protein 104 |
| chr3_-_194207388 | 0.07 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chr17_-_7297519 | 0.07 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr17_+_72428218 | 0.07 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr18_-_53255766 | 0.07 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr19_-_663171 | 0.07 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr7_+_128502895 | 0.07 |
ENST00000492758.1
|
ATP6V1F
|
ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F |
| chr22_+_31090793 | 0.07 |
ENST00000332585.6
ENST00000382310.3 ENST00000446658.2 |
OSBP2
|
oxysterol binding protein 2 |
| chr7_-_5569588 | 0.07 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr5_-_176923846 | 0.07 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr11_+_66045634 | 0.07 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr1_-_35395178 | 0.07 |
ENST00000373347.1
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr11_-_66103932 | 0.07 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr1_+_155051379 | 0.07 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr19_+_35634146 | 0.07 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr16_+_3070356 | 0.07 |
ENST00000341627.5
ENST00000575124.1 ENST00000575836.1 |
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr5_+_176853702 | 0.07 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr2_-_38830030 | 0.07 |
ENST00000410076.1
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr1_-_156786634 | 0.07 |
ENST00000392306.2
ENST00000368199.3 |
SH2D2A
|
SH2 domain containing 2A |
| chr5_+_176853669 | 0.07 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr16_-_66959429 | 0.07 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr11_-_64512273 | 0.06 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr16_-_2185899 | 0.06 |
ENST00000262304.4
ENST00000423118.1 |
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr16_+_29823427 | 0.06 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr17_-_17494972 | 0.06 |
ENST00000435340.2
ENST00000255389.5 ENST00000395781.2 |
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr11_-_118966167 | 0.06 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr1_+_156698708 | 0.06 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr11_-_64014379 | 0.06 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_+_156698234 | 0.06 |
ENST00000368218.4
ENST00000368216.4 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr17_-_41623716 | 0.06 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr19_+_14544099 | 0.06 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr8_+_144821557 | 0.06 |
ENST00000534398.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr1_+_35734616 | 0.06 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr16_-_29910365 | 0.06 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr17_+_75369400 | 0.06 |
ENST00000590059.1
|
SEPT9
|
septin 9 |
| chr6_+_12012536 | 0.06 |
ENST00000379388.2
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr15_-_71146347 | 0.06 |
ENST00000559140.2
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr9_-_130742792 | 0.06 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr16_+_28834303 | 0.06 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chr5_+_149109825 | 0.06 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr22_+_38597889 | 0.06 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr7_+_27282319 | 0.06 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr8_-_145911188 | 0.06 |
ENST00000540274.1
|
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr7_-_559853 | 0.06 |
ENST00000405692.2
|
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr17_-_79995553 | 0.06 |
ENST00000581584.1
ENST00000577712.1 ENST00000582900.1 ENST00000579155.1 ENST00000306869.2 |
DCXR
|
dicarbonyl/L-xylulose reductase |
| chr18_+_3449695 | 0.06 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr21_+_44394742 | 0.06 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr12_+_107349606 | 0.06 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chrX_+_153237740 | 0.06 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chr22_-_24181174 | 0.06 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr19_-_9929708 | 0.06 |
ENST00000247977.4
ENST00000590277.1 ENST00000588922.1 ENST00000589626.1 ENST00000592067.1 ENST00000586469.1 |
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr19_+_676385 | 0.06 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr12_-_49582593 | 0.06 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr3_+_75721428 | 0.05 |
ENST00000463183.1
|
LINC00960
|
long intergenic non-protein coding RNA 960 |
| chr6_+_134274322 | 0.05 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr16_+_2933209 | 0.05 |
ENST00000293981.6
|
FLYWCH2
|
FLYWCH family member 2 |
| chr1_-_15850676 | 0.05 |
ENST00000440484.1
ENST00000333868.5 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr19_+_54412517 | 0.05 |
ENST00000391767.1
|
CACNG7
|
calcium channel, voltage-dependent, gamma subunit 7 |
| chr11_+_46403194 | 0.05 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr14_-_23770683 | 0.05 |
ENST00000561437.1
ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr8_+_128747661 | 0.05 |
ENST00000259523.6
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr11_+_46402583 | 0.05 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr7_-_100860851 | 0.05 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr19_-_36505098 | 0.05 |
ENST00000252984.7
ENST00000486389.1 ENST00000378875.3 ENST00000485128.1 |
ALKBH6
|
alkB, alkylation repair homolog 6 (E. coli) |
| chrX_+_37545012 | 0.05 |
ENST00000378616.3
|
XK
|
X-linked Kx blood group (McLeod syndrome) |
| chr11_-_64511575 | 0.05 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr11_+_67070919 | 0.05 |
ENST00000308127.4
ENST00000308298.7 |
SSH3
|
slingshot protein phosphatase 3 |
| chr8_-_145331153 | 0.05 |
ENST00000377412.4
|
KM-PA-2
|
KM-PA-2 protein; Uncharacterized protein |
| chr12_+_56661033 | 0.05 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr5_+_63461642 | 0.05 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr3_+_88108381 | 0.05 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr1_-_182573514 | 0.05 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr11_-_123525289 | 0.05 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr19_-_40724246 | 0.05 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr3_-_45267760 | 0.05 |
ENST00000503771.1
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene) |
| chr2_+_191208601 | 0.05 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr1_-_6052463 | 0.05 |
ENST00000378156.4
|
NPHP4
|
nephronophthisis 4 |
| chr5_+_149865838 | 0.05 |
ENST00000519157.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr11_+_66610883 | 0.05 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr4_+_109571800 | 0.05 |
ENST00000512478.2
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr1_+_6673745 | 0.05 |
ENST00000377648.4
|
PHF13
|
PHD finger protein 13 |
| chr9_+_74764340 | 0.05 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr7_+_43966018 | 0.05 |
ENST00000222402.3
ENST00000446008.1 ENST00000394798.4 |
UBE2D4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr5_+_139028510 | 0.05 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr17_-_18945798 | 0.05 |
ENST00000395635.1
|
GRAP
|
GRB2-related adaptor protein |
| chr11_-_66104237 | 0.05 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr17_-_72772462 | 0.05 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr9_-_34637718 | 0.05 |
ENST00000378892.1
ENST00000277010.4 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr16_+_28986085 | 0.05 |
ENST00000565975.1
ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr20_+_48429233 | 0.05 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr19_-_55691614 | 0.05 |
ENST00000592470.1
ENST00000354308.3 |
SYT5
|
synaptotagmin V |
| chr17_-_4710288 | 0.05 |
ENST00000571067.1
|
RP11-81A22.5
|
RP11-81A22.5 |
| chr19_+_7985880 | 0.05 |
ENST00000597584.1
|
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr20_-_62258394 | 0.05 |
ENST00000370077.1
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr19_-_55919087 | 0.05 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr8_-_145060593 | 0.05 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chrX_+_16964794 | 0.05 |
ENST00000357277.3
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr11_-_77899634 | 0.05 |
ENST00000526208.1
ENST00000529350.1 ENST00000530018.1 ENST00000528776.1 |
KCTD21
|
potassium channel tetramerization domain containing 21 |
| chr20_+_36531499 | 0.05 |
ENST00000373458.3
ENST00000373461.4 ENST00000373459.4 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr7_+_116593953 | 0.05 |
ENST00000397750.3
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr22_-_24110063 | 0.05 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr19_-_45953983 | 0.05 |
ENST00000592083.1
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr1_+_32671236 | 0.05 |
ENST00000537469.1
ENST00000291358.6 |
IQCC
|
IQ motif containing C |
| chr19_+_1275917 | 0.05 |
ENST00000469144.1
|
C19orf24
|
chromosome 19 open reading frame 24 |
| chr7_+_100199800 | 0.05 |
ENST00000223061.5
|
PCOLCE
|
procollagen C-endopeptidase enhancer |
| chr12_-_53625958 | 0.05 |
ENST00000327550.3
ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG
|
retinoic acid receptor, gamma |
| chr12_+_1099675 | 0.05 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr16_+_2563871 | 0.05 |
ENST00000330398.4
ENST00000568562.1 ENST00000569317.1 |
ATP6V0C
ATP6C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c Uncharacterized protein |
| chr19_+_39897453 | 0.05 |
ENST00000597629.1
ENST00000248673.3 ENST00000594045.1 ENST00000594442.1 |
ZFP36
|
ZFP36 ring finger protein |
| chr3_-_49907323 | 0.05 |
ENST00000296471.7
ENST00000488336.1 ENST00000467248.1 ENST00000466940.1 ENST00000463537.1 ENST00000480398.2 |
CAMKV
|
CaM kinase-like vesicle-associated |
| chr19_+_19303572 | 0.05 |
ENST00000407360.3
ENST00000540981.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
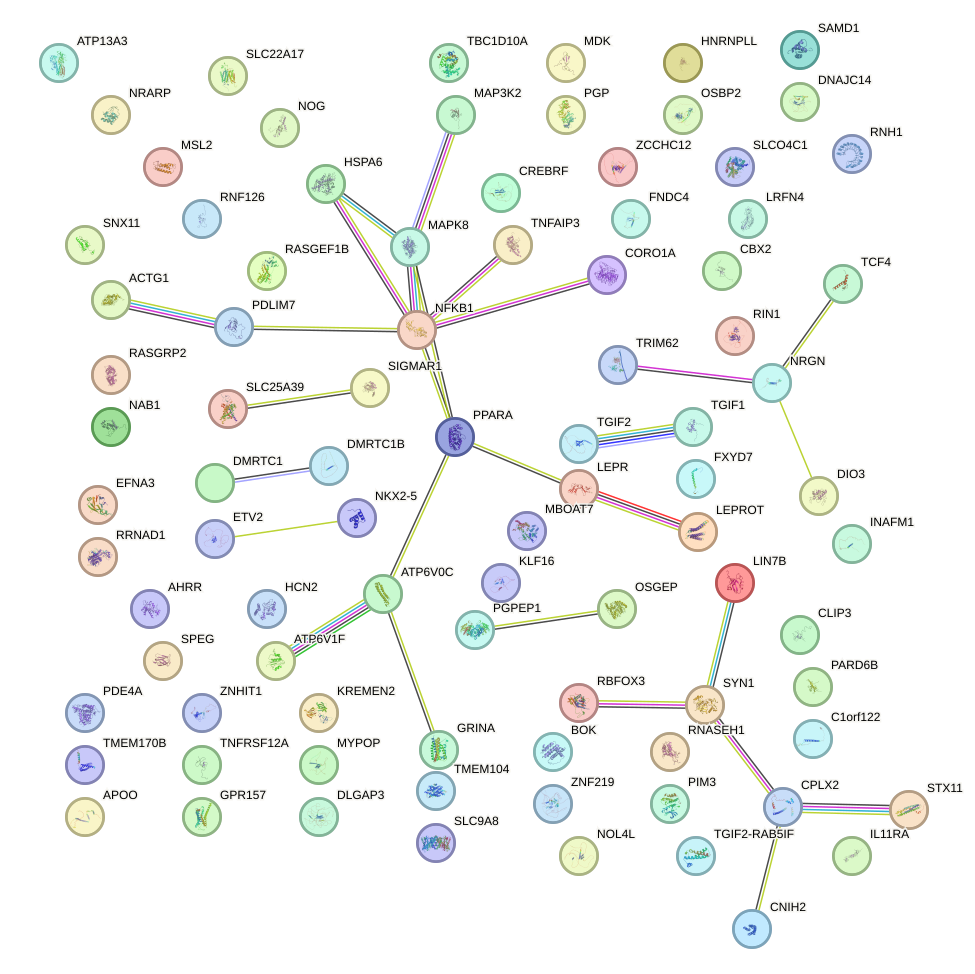
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR1_EGR4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003168 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:2000349 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.0 | GO:0071505 | response to mycophenolic acid(GO:0071505) cellular response to mycophenolic acid(GO:0071506) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.2 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.3 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0090108 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of pancreatic juice secretion(GO:0090187) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.0 | 0.1 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0061341 | chemoattraction of serotonergic neuron axon(GO:0036517) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.0 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.0 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.1 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.0 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.0 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.0 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0033962 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.0 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |