|
chr6_+_27833034
Show fit
|
1.32 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al
|
|
chr6_+_27782788
Show fit
|
0.76 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm
|
|
chr6_+_24775641
Show fit
|
0.73 |
ENST00000378054.2
ENST00000476555.1
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr1_-_91487013
Show fit
|
0.64 |
ENST00000347275.5
ENST00000370440.1
|
ZNF644
|
zinc finger protein 644
|
|
chr11_-_19263145
Show fit
|
0.63 |
ENST00000532666.1
ENST00000527884.1
|
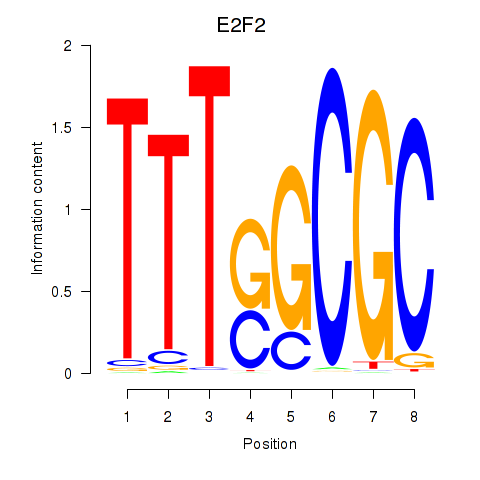
E2F8
|
E2F transcription factor 8
|
|
chr11_-_95522907
Show fit
|
0.59 |
ENST00000358780.5
ENST00000542135.1
|
FAM76B
|
family with sequence similarity 76, member B
|
|
chr15_-_71184724
Show fit
|
0.56 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10
|
|
chr14_+_36295638
Show fit
|
0.56 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like
|
|
chr11_-_95522639
Show fit
|
0.52 |
ENST00000536839.1
|
FAM76B
|
family with sequence similarity 76, member B
|
|
chr18_+_19192228
Show fit
|
0.48 |
ENST00000300413.5
ENST00000579618.1
ENST00000582475.1
|
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa
|
|
chr1_+_228645796
Show fit
|
0.48 |
ENST00000369160.2
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chrX_-_152736013
Show fit
|
0.48 |
ENST00000330912.2
ENST00000338525.2
ENST00000334497.2
ENST00000370232.1
ENST00000370212.3
ENST00000370211.4
|
TREX2
HAUS7
|
three prime repair exonuclease 2
HAUS augmin-like complex, subunit 7
|
|
chr11_-_57102947
Show fit
|
0.44 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1
|
|
chr1_-_26232522
Show fit
|
0.44 |
ENST00000399728.1
|
STMN1
|
stathmin 1
|
|
chr10_+_62538248
Show fit
|
0.41 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr22_+_20105259
Show fit
|
0.41 |
ENST00000416427.1
ENST00000421656.1
ENST00000423859.1
ENST00000418705.2
|
RANBP1
|
RAN binding protein 1
|
|
chr19_+_58694742
Show fit
|
0.41 |
ENST00000597528.1
ENST00000594839.1
|
ZNF274
|
zinc finger protein 274
|
|
chr7_+_26241325
Show fit
|
0.39 |
ENST00000456948.1
ENST00000409747.1
|
CBX3
|
chromobox homolog 3
|
|
chr5_-_79950371
Show fit
|
0.38 |
ENST00000511032.1
ENST00000504396.1
ENST00000505337.1
|
DHFR
|
dihydrofolate reductase
|
|
chr19_+_8455200
Show fit
|
0.38 |
ENST00000601897.1
ENST00000594216.1
|
RAB11B
|
RAB11B, member RAS oncogene family
|
|
chr3_+_10857885
Show fit
|
0.38 |
ENST00000254488.2
ENST00000454147.1
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11
|
|
chr22_+_20105012
Show fit
|
0.38 |
ENST00000331821.3
ENST00000411892.1
|
RANBP1
|
RAN binding protein 1
|
|
chr12_-_12849073
Show fit
|
0.38 |
ENST00000332427.2
ENST00000540796.1
|
GPR19
|
G protein-coupled receptor 19
|
|
chrX_-_45710920
Show fit
|
0.37 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3
|
|
chr19_-_10305302
Show fit
|
0.37 |
ENST00000592054.1
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1
|
|
chr2_-_55277692
Show fit
|
0.37 |
ENST00000394611.2
|
RTN4
|
reticulon 4
|
|
chr5_-_36152031
Show fit
|
0.37 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2
|
|
chr10_+_103912137
Show fit
|
0.37 |
ENST00000603742.1
ENST00000488254.2
ENST00000461421.1
ENST00000476468.1
ENST00000370007.5
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr2_-_203103281
Show fit
|
0.36 |
ENST00000392244.3
ENST00000409181.1
ENST00000409712.1
ENST00000409498.2
ENST00000409368.1
ENST00000392245.1
ENST00000392246.2
|
SUMO1
|
small ubiquitin-like modifier 1
|
|
chr6_-_132272504
Show fit
|
0.36 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor
|
|
chr1_-_36235529
Show fit
|
0.35 |
ENST00000318121.3
ENST00000373220.3
ENST00000520551.1
|
CLSPN
|
claspin
|
|
chr1_+_212782012
Show fit
|
0.35 |
ENST00000341491.4
ENST00000366985.1
|
ATF3
|
activating transcription factor 3
|
|
chr7_+_26241310
Show fit
|
0.34 |
ENST00000396386.2
|
CBX3
|
chromobox homolog 3
|
|
chr17_-_77179487
Show fit
|
0.34 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr19_+_58694396
Show fit
|
0.34 |
ENST00000326804.4
ENST00000345813.3
ENST00000424679.2
|
ZNF274
|
zinc finger protein 274
|
|
chr1_-_91487770
Show fit
|
0.33 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644
|
|
chr16_-_2827128
Show fit
|
0.33 |
ENST00000494946.2
ENST00000409477.1
ENST00000572954.1
ENST00000262306.7
ENST00000409906.4
|
TCEB2
|
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B)
|
|
chr2_-_136633940
Show fit
|
0.33 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6
|
|
chr17_-_17184605
Show fit
|
0.31 |
ENST00000268717.5
|
COPS3
|
COP9 signalosome subunit 3
|
|
chr17_+_29158962
Show fit
|
0.31 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5
|
|
chr8_-_94928861
Show fit
|
0.31 |
ENST00000607097.1
|
MIR378D2
|
microRNA 378d-2
|
|
chr22_+_20104947
Show fit
|
0.30 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1
|
|
chr2_-_55277436
Show fit
|
0.30 |
ENST00000354474.6
|
RTN4
|
reticulon 4
|
|
chr20_+_5931275
Show fit
|
0.29 |
ENST00000378896.3
ENST00000378883.1
|
MCM8
|
minichromosome maintenance complex component 8
|
|
chr6_-_27114577
Show fit
|
0.29 |
ENST00000356950.1
ENST00000396891.4
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr1_-_212208842
Show fit
|
0.29 |
ENST00000366992.3
ENST00000366993.3
ENST00000440600.2
ENST00000366994.3
|
INTS7
|
integrator complex subunit 7
|
|
chr2_+_17935119
Show fit
|
0.28 |
ENST00000317402.7
|
GEN1
|
GEN1 Holliday junction 5' flap endonuclease
|
|
chr2_+_48010312
Show fit
|
0.28 |
ENST00000540021.1
|
MSH6
|
mutS homolog 6
|
|
chr1_-_91487806
Show fit
|
0.28 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644
|
|
chrX_-_135849484
Show fit
|
0.27 |
ENST00000370620.1
ENST00000535227.1
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr13_+_114238997
Show fit
|
0.27 |
ENST00000538138.1
ENST00000375370.5
|
TFDP1
|
transcription factor Dp-1
|
|
chr8_-_124408652
Show fit
|
0.27 |
ENST00000287394.5
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chr6_+_27775899
Show fit
|
0.26 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai
|
|
chr11_-_95523500
Show fit
|
0.26 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B
|
|
chr11_-_19262486
Show fit
|
0.25 |
ENST00000250024.4
|
E2F8
|
E2F transcription factor 8
|
|
chr6_+_31633011
Show fit
|
0.25 |
ENST00000375885.4
|
CSNK2B
|
casein kinase 2, beta polypeptide
|
|
chr12_-_125398850
Show fit
|
0.25 |
ENST00000535859.1
ENST00000546271.1
ENST00000540700.1
ENST00000546120.1
|
UBC
|
ubiquitin C
|
|
chr10_+_112327425
Show fit
|
0.25 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr1_-_36235559
Show fit
|
0.25 |
ENST00000251195.5
|
CLSPN
|
claspin
|
|
chr17_+_42733803
Show fit
|
0.25 |
ENST00000409122.2
|
C17orf104
|
chromosome 17 open reading frame 104
|
|
chr6_+_24775153
Show fit
|
0.25 |
ENST00000356509.3
ENST00000230056.3
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr1_+_6845497
Show fit
|
0.24 |
ENST00000473578.1
ENST00000557126.1
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr15_+_40987327
Show fit
|
0.24 |
ENST00000423169.2
ENST00000267868.3
ENST00000557850.1
ENST00000532743.1
ENST00000382643.3
|
RAD51
|
RAD51 recombinase
|
|
chr15_+_40987661
Show fit
|
0.24 |
ENST00000526763.1
|
RAD51
|
RAD51 recombinase
|
|
chr10_+_103911926
Show fit
|
0.24 |
ENST00000605788.1
ENST00000405356.1
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr3_-_44803141
Show fit
|
0.24 |
ENST00000296121.4
|
KIAA1143
|
KIAA1143
|
|
chr1_-_47779762
Show fit
|
0.24 |
ENST00000371877.3
ENST00000360380.3
ENST00000337817.5
ENST00000447475.2
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr6_-_17706618
Show fit
|
0.24 |
ENST00000262077.2
ENST00000537253.1
|
NUP153
|
nucleoporin 153kDa
|
|
chrX_+_123095546
Show fit
|
0.23 |
ENST00000371157.3
ENST00000371145.3
ENST00000371144.3
|
STAG2
|
stromal antigen 2
|
|
chr20_+_5931497
Show fit
|
0.23 |
ENST00000378886.2
ENST00000265187.4
|
MCM8
|
minichromosome maintenance complex component 8
|
|
chr3_-_10028366
Show fit
|
0.23 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3
|
|
chr7_-_6048702
Show fit
|
0.23 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae)
|
|
chr2_-_61108449
Show fit
|
0.23 |
ENST00000439412.1
ENST00000452343.1
|
AC010733.4
|
AC010733.4
|
|
chr6_+_116575329
Show fit
|
0.22 |
ENST00000430252.2
ENST00000540275.1
ENST00000448740.2
|
DSE
RP3-486I3.7
|
dermatan sulfate epimerase
RP3-486I3.7
|
|
chr8_+_94929077
Show fit
|
0.22 |
ENST00000297598.4
ENST00000520614.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr4_+_55524085
Show fit
|
0.22 |
ENST00000412167.2
ENST00000288135.5
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr7_-_105516923
Show fit
|
0.22 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr22_+_35653445
Show fit
|
0.22 |
ENST00000420166.1
ENST00000444518.2
ENST00000455359.1
ENST00000216106.5
|
HMGXB4
|
HMG box domain containing 4
|
|
chr4_+_71859156
Show fit
|
0.21 |
ENST00000286648.5
ENST00000504730.1
ENST00000504952.1
|
DCK
|
deoxycytidine kinase
|
|
chr13_-_26625169
Show fit
|
0.21 |
ENST00000319420.3
|
SHISA2
|
shisa family member 2
|
|
chr2_+_48010221
Show fit
|
0.21 |
ENST00000234420.5
|
MSH6
|
mutS homolog 6
|
|
chr2_-_203103185
Show fit
|
0.21 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1
|
|
chrX_+_21959108
Show fit
|
0.21 |
ENST00000457085.1
|
SMS
|
spermine synthase
|
|
chr2_-_232329186
Show fit
|
0.20 |
ENST00000322723.4
|
NCL
|
nucleolin
|
|
chr20_-_5931109
Show fit
|
0.20 |
ENST00000203001.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae)
|
|
chr13_+_32889605
Show fit
|
0.20 |
ENST00000380152.3
ENST00000544455.1
ENST00000530893.2
|
BRCA2
|
breast cancer 2, early onset
|
|
chr1_-_26233423
Show fit
|
0.20 |
ENST00000357865.2
|
STMN1
|
stathmin 1
|
|
chr9_-_123639304
Show fit
|
0.20 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19
|
|
chr19_+_35521699
Show fit
|
0.20 |
ENST00000415950.3
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit
|
|
chr8_+_94929168
Show fit
|
0.20 |
ENST00000518107.1
ENST00000396200.3
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr1_-_26232951
Show fit
|
0.20 |
ENST00000426559.2
ENST00000455785.2
|
STMN1
|
stathmin 1
|
|
chr17_+_30264014
Show fit
|
0.20 |
ENST00000322652.5
ENST00000580398.1
|
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit
|
|
chr1_+_222885884
Show fit
|
0.20 |
ENST00000340934.5
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr5_+_138629389
Show fit
|
0.20 |
ENST00000504045.1
ENST00000504311.1
ENST00000502499.1
|
MATR3
|
matrin 3
|
|
chr15_+_100106244
Show fit
|
0.19 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr2_+_174219548
Show fit
|
0.19 |
ENST00000347703.3
ENST00000392567.2
ENST00000306721.3
ENST00000410101.3
ENST00000410019.3
|
CDCA7
|
cell division cycle associated 7
|
|
chr2_-_17935027
Show fit
|
0.19 |
ENST00000446852.1
|
SMC6
|
structural maintenance of chromosomes 6
|
|
chr11_-_64739358
Show fit
|
0.19 |
ENST00000301896.5
ENST00000530444.1
|
C11orf85
|
chromosome 11 open reading frame 85
|
|
chr7_+_116593433
Show fit
|
0.19 |
ENST00000323984.3
ENST00000393449.1
|
ST7
|
suppression of tumorigenicity 7
|
|
chr8_+_94929273
Show fit
|
0.19 |
ENST00000518573.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr2_+_238707388
Show fit
|
0.19 |
ENST00000316997.4
|
RBM44
|
RNA binding motif protein 44
|
|
chr6_+_135502408
Show fit
|
0.19 |
ENST00000341911.5
ENST00000442647.2
ENST00000316528.8
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog
|
|
chr5_+_43121607
Show fit
|
0.18 |
ENST00000509156.1
ENST00000508259.1
ENST00000306938.4
ENST00000399534.1
|
ZNF131
|
zinc finger protein 131
|
|
chr19_+_36705504
Show fit
|
0.18 |
ENST00000456324.1
|
ZNF146
|
zinc finger protein 146
|
|
chrX_+_53449887
Show fit
|
0.18 |
ENST00000375327.3
|
RIBC1
|
RIB43A domain with coiled-coils 1
|
|
chr2_-_17935059
Show fit
|
0.18 |
ENST00000448223.2
ENST00000381272.4
ENST00000351948.4
|
SMC6
|
structural maintenance of chromosomes 6
|
|
chr19_-_36705547
Show fit
|
0.18 |
ENST00000304116.5
|
ZNF565
|
zinc finger protein 565
|
|
chr19_+_36706024
Show fit
|
0.18 |
ENST00000443387.2
|
ZNF146
|
zinc finger protein 146
|
|
chr12_-_125399573
Show fit
|
0.18 |
ENST00000339647.5
|
UBC
|
ubiquitin C
|
|
chr3_-_48229846
Show fit
|
0.18 |
ENST00000302506.3
ENST00000351231.3
ENST00000437972.1
|
CDC25A
|
cell division cycle 25A
|
|
chr7_+_116593536
Show fit
|
0.18 |
ENST00000417919.1
|
ST7
|
suppression of tumorigenicity 7
|
|
chr1_+_100598691
Show fit
|
0.18 |
ENST00000370143.1
ENST00000370141.2
|
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae)
|
|
chr3_-_136471204
Show fit
|
0.18 |
ENST00000480733.1
ENST00000383202.2
ENST00000236698.5
ENST00000434713.2
|
STAG1
|
stromal antigen 1
|
|
chr11_+_13299186
Show fit
|
0.17 |
ENST00000527998.1
ENST00000396441.3
ENST00000533520.1
ENST00000529825.1
ENST00000389707.4
ENST00000401424.1
ENST00000529388.1
ENST00000530357.1
ENST00000403290.1
ENST00000361003.4
ENST00000389708.3
ENST00000403510.3
ENST00000482049.1
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr5_+_43121698
Show fit
|
0.17 |
ENST00000505606.2
ENST00000509634.1
ENST00000509341.1
|
ZNF131
|
zinc finger protein 131
|
|
chr7_+_116593568
Show fit
|
0.17 |
ENST00000446490.1
|
ST7
|
suppression of tumorigenicity 7
|
|
chr13_+_115079949
Show fit
|
0.17 |
ENST00000361283.1
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1
|
|
chr14_+_35452104
Show fit
|
0.17 |
ENST00000216774.6
ENST00000546080.1
|
SRP54
|
signal recognition particle 54kDa
|
|
chrX_+_46696372
Show fit
|
0.17 |
ENST00000218340.3
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive)
|
|
chr5_-_122759032
Show fit
|
0.17 |
ENST00000510582.3
ENST00000328236.5
ENST00000306481.6
ENST00000508442.2
ENST00000395431.2
|
CEP120
|
centrosomal protein 120kDa
|
|
chr11_+_9595180
Show fit
|
0.17 |
ENST00000450114.2
|
WEE1
|
WEE1 G2 checkpoint kinase
|
|
chr8_+_128748308
Show fit
|
0.17 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog
|
|
chr5_-_79950775
Show fit
|
0.17 |
ENST00000439211.2
|
DHFR
|
dihydrofolate reductase
|
|
chr6_-_52149475
Show fit
|
0.16 |
ENST00000419835.2
ENST00000229854.7
ENST00000596288.1
|
MCM3
|
minichromosome maintenance complex component 3
|
|
chr12_-_77459306
Show fit
|
0.16 |
ENST00000547316.1
ENST00000416496.2
ENST00000550669.1
ENST00000322886.7
|
E2F7
|
E2F transcription factor 7
|
|
chr11_-_59436453
Show fit
|
0.16 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast)
|
|
chr9_-_99180597
Show fit
|
0.16 |
ENST00000375256.4
|
ZNF367
|
zinc finger protein 367
|
|
chr20_-_35724388
Show fit
|
0.16 |
ENST00000344359.3
ENST00000373664.3
|
RBL1
|
retinoblastoma-like 1 (p107)
|
|
chr1_-_222885770
Show fit
|
0.16 |
ENST00000355727.2
ENST00000340020.6
|
AIDA
|
axin interactor, dorsalization associated
|
|
chr6_-_160210604
Show fit
|
0.16 |
ENST00000420894.2
ENST00000539756.1
ENST00000544255.1
|
TCP1
|
t-complex 1
|
|
chr3_+_14219858
Show fit
|
0.16 |
ENST00000306024.3
|
LSM3
|
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr8_-_120868078
Show fit
|
0.16 |
ENST00000313655.4
|
DSCC1
|
DNA replication and sister chromatid cohesion 1
|
|
chr7_+_90225796
Show fit
|
0.16 |
ENST00000380050.3
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr15_+_71185148
Show fit
|
0.16 |
ENST00000443425.2
ENST00000560755.1
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr17_+_42733730
Show fit
|
0.15 |
ENST00000359945.3
ENST00000425535.1
|
C17orf104
|
chromosome 17 open reading frame 104
|
|
chr7_-_148581251
Show fit
|
0.15 |
ENST00000478654.1
ENST00000460911.1
ENST00000350995.2
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr10_+_96305610
Show fit
|
0.15 |
ENST00000371332.4
ENST00000239026.6
|
HELLS
|
helicase, lymphoid-specific
|
|
chr14_+_51706886
Show fit
|
0.15 |
ENST00000457354.2
|
TMX1
|
thioredoxin-related transmembrane protein 1
|
|
chr14_+_36295504
Show fit
|
0.15 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like
|
|
chr3_+_41240925
Show fit
|
0.15 |
ENST00000396183.3
ENST00000349496.5
ENST00000453024.1
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa
|
|
chr2_-_219433014
Show fit
|
0.15 |
ENST00000418019.1
ENST00000454775.1
ENST00000338465.5
ENST00000415516.1
ENST00000258399.3
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr7_-_559853
Show fit
|
0.14 |
ENST00000405692.2
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr17_-_73781567
Show fit
|
0.14 |
ENST00000586607.1
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr7_+_94285637
Show fit
|
0.14 |
ENST00000482108.1
ENST00000488574.1
|
PEG10
|
paternally expressed 10
|
|
chr2_+_48541776
Show fit
|
0.14 |
ENST00000413569.1
ENST00000340553.3
|
FOXN2
|
forkhead box N2
|
|
chr5_+_36876833
Show fit
|
0.14 |
ENST00000282516.8
ENST00000448238.2
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr6_-_144329531
Show fit
|
0.14 |
ENST00000429150.1
ENST00000392309.1
ENST00000416623.1
ENST00000392307.1
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr7_+_116593292
Show fit
|
0.14 |
ENST00000393446.2
ENST00000265437.5
ENST00000393451.3
|
ST7
|
suppression of tumorigenicity 7
|
|
chr7_+_120591170
Show fit
|
0.14 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3
|
|
chr7_-_148581360
Show fit
|
0.14 |
ENST00000320356.2
ENST00000541220.1
ENST00000483967.1
ENST00000536783.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr1_+_222988464
Show fit
|
0.14 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3
|
|
chr7_+_26240776
Show fit
|
0.14 |
ENST00000337620.4
|
CBX3
|
chromobox homolog 3
|
|
chr15_+_100106126
Show fit
|
0.14 |
ENST00000558812.1
ENST00000338042.6
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr8_-_67525473
Show fit
|
0.14 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1
|
|
chr20_-_5931051
Show fit
|
0.14 |
ENST00000453074.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae)
|
|
chr2_-_96700664
Show fit
|
0.14 |
ENST00000359548.4
ENST00000377137.3
ENST00000439254.1
ENST00000453542.1
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial
|
|
chr4_+_72052964
Show fit
|
0.14 |
ENST00000264485.5
ENST00000425175.1
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4
|
|
chrX_+_118708517
Show fit
|
0.14 |
ENST00000346330.3
|
UBE2A
|
ubiquitin-conjugating enzyme E2A
|
|
chr13_-_73356009
Show fit
|
0.14 |
ENST00000377780.4
ENST00000377767.4
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr1_-_23670817
Show fit
|
0.13 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R
|
|
chr10_-_13390021
Show fit
|
0.13 |
ENST00000537130.1
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chrX_+_118708493
Show fit
|
0.13 |
ENST00000371558.2
|
UBE2A
|
ubiquitin-conjugating enzyme E2A
|
|
chr17_-_56065540
Show fit
|
0.13 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chrX_+_131157322
Show fit
|
0.13 |
ENST00000481105.1
ENST00000354719.6
ENST00000394335.2
|
MST4
|
Serine/threonine-protein kinase MST4
|
|
chrX_+_131157290
Show fit
|
0.13 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4
|
|
chrX_+_24711997
Show fit
|
0.13 |
ENST00000379068.3
ENST00000379059.3
|
POLA1
|
polymerase (DNA directed), alpha 1, catalytic subunit
|
|
chr20_+_25388293
Show fit
|
0.13 |
ENST00000262460.4
ENST00000429262.2
|
GINS1
|
GINS complex subunit 1 (Psf1 homolog)
|
|
chr5_+_138629337
Show fit
|
0.13 |
ENST00000394805.3
ENST00000512876.1
ENST00000513678.1
|
MATR3
|
matrin 3
|
|
chr14_+_35452169
Show fit
|
0.13 |
ENST00000555557.1
|
SRP54
|
signal recognition particle 54kDa
|
|
chr1_-_243418344
Show fit
|
0.13 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa
|
|
chr11_-_77705687
Show fit
|
0.13 |
ENST00000529807.1
ENST00000527522.1
ENST00000534064.1
|
INTS4
|
integrator complex subunit 4
|
|
chr1_-_23670813
Show fit
|
0.13 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R
|
|
chr7_+_120590803
Show fit
|
0.13 |
ENST00000315870.5
ENST00000339121.5
ENST00000445699.1
|
ING3
|
inhibitor of growth family, member 3
|
|
chrX_+_70798261
Show fit
|
0.13 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing
|
|
chr17_+_7477040
Show fit
|
0.13 |
ENST00000581384.1
ENST00000577929.1
|
EIF4A1
|
eukaryotic translation initiation factor 4A1
|
|
chr1_-_229694406
Show fit
|
0.12 |
ENST00000344517.4
|
ABCB10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10
|
|
chrX_-_53449593
Show fit
|
0.12 |
ENST00000375340.6
ENST00000322213.4
|
SMC1A
|
structural maintenance of chromosomes 1A
|
|
chr14_-_50154921
Show fit
|
0.12 |
ENST00000553805.2
ENST00000554396.1
ENST00000216367.5
ENST00000539565.2
|
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit
|
|
chr6_-_160210715
Show fit
|
0.12 |
ENST00000392168.2
ENST00000321394.7
|
TCP1
|
t-complex 1
|
|
chr7_+_94139105
Show fit
|
0.12 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1
|
|
chr4_+_72053017
Show fit
|
0.12 |
ENST00000351898.6
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4
|
|
chr2_+_219433588
Show fit
|
0.12 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr3_+_44803209
Show fit
|
0.12 |
ENST00000326047.4
|
KIF15
|
kinesin family member 15
|
|
chr22_+_42394780
Show fit
|
0.12 |
ENST00000328823.9
|
WBP2NL
|
WBP2 N-terminal like
|
|
chr17_+_58677539
Show fit
|
0.12 |
ENST00000305921.3
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr15_+_80987617
Show fit
|
0.12 |
ENST00000258884.4
ENST00000558464.1
|
ABHD17C
|
abhydrolase domain containing 17C
|
|
chr5_+_93954358
Show fit
|
0.12 |
ENST00000504099.1
|
ANKRD32
|
ankyrin repeat domain 32
|
|
chr1_-_243418621
Show fit
|
0.12 |
ENST00000366544.1
ENST00000366543.1
|
CEP170
|
centrosomal protein 170kDa
|
|
chrX_-_131623982
Show fit
|
0.11 |
ENST00000370844.1
|
MBNL3
|
muscleblind-like splicing regulator 3
|
|
chr3_-_133380731
Show fit
|
0.11 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1
|
|
chr12_-_48499591
Show fit
|
0.11 |
ENST00000551330.1
ENST00000004980.5
ENST00000339976.6
ENST00000448372.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1
|
|
chr12_-_122238464
Show fit
|
0.11 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia)
|
|
chr11_+_74303612
Show fit
|
0.11 |
ENST00000527458.1
ENST00000532497.1
ENST00000530511.1
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit
|
|
chr12_-_81331460
Show fit
|
0.11 |
ENST00000549417.1
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr4_-_170679024
Show fit
|
0.11 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27
|
|
chr12_-_133263893
Show fit
|
0.11 |
ENST00000535270.1
ENST00000320574.5
|
POLE
|
polymerase (DNA directed), epsilon, catalytic subunit
|
|
chr6_-_27100529
Show fit
|
0.11 |
ENST00000607124.1
ENST00000339812.2
ENST00000541790.1
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr7_-_99699538
Show fit
|
0.11 |
ENST00000343023.6
ENST00000303887.5
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr4_-_130014532
Show fit
|
0.11 |
ENST00000506368.1
ENST00000439369.2
ENST00000503215.1
|
SCLT1
|
sodium channel and clathrin linker 1
|
|
chr14_-_69864993
Show fit
|
0.10 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila)
|