Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
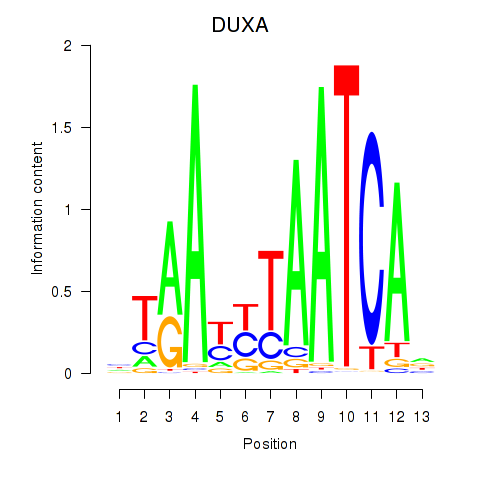
Results for DUXA
Z-value: 1.46
Transcription factors associated with DUXA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DUXA
|
ENSG00000258873.2 | double homeobox A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DUXA | hg19_v2_chr19_-_57678811_57678811 | 0.54 | 4.6e-01 | Click! |
Activity profile of DUXA motif
Sorted Z-values of DUXA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91546926 | 1.15 |
ENST00000550758.1
|
DCN
|
decorin |
| chr1_-_168464875 | 0.90 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr3_+_159943362 | 0.81 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr4_+_156775910 | 0.72 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr7_+_30589829 | 0.72 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr1_+_95616933 | 0.70 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr6_+_28092338 | 0.62 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr16_-_57219926 | 0.59 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr8_-_86575726 | 0.58 |
ENST00000379010.2
|
REXO1L1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1, pseudogene |
| chr2_+_242913327 | 0.56 |
ENST00000426962.1
|
AC093642.3
|
AC093642.3 |
| chr2_-_69180083 | 0.54 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr3_+_118892411 | 0.50 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr8_+_92114060 | 0.50 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr16_+_53412368 | 0.50 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr6_+_86195088 | 0.48 |
ENST00000437581.1
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr2_+_86333340 | 0.48 |
ENST00000409783.2
ENST00000409277.3 |
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr16_-_57219721 | 0.47 |
ENST00000562406.1
ENST00000568671.1 ENST00000567044.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr14_+_21566980 | 0.45 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr10_-_112064665 | 0.44 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr10_-_112255945 | 0.44 |
ENST00000609514.1
ENST00000607952.1 |
RP11-525A16.4
|
RP11-525A16.4 |
| chrX_-_55187531 | 0.43 |
ENST00000489298.1
ENST00000477847.2 |
FAM104B
|
family with sequence similarity 104, member B |
| chr11_-_95523500 | 0.43 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr7_+_141490017 | 0.40 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr1_+_40997233 | 0.40 |
ENST00000372699.3
ENST00000372697.3 ENST00000372696.3 |
ZNF684
|
zinc finger protein 684 |
| chr11_+_10772847 | 0.40 |
ENST00000524523.1
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr2_-_114461655 | 0.40 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr6_+_168434678 | 0.40 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr12_-_75784669 | 0.39 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr15_+_71228826 | 0.39 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr11_+_10772534 | 0.39 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr4_+_77941685 | 0.37 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chrX_-_46759138 | 0.37 |
ENST00000377879.3
|
CXorf31
|
chromosome X open reading frame 31 |
| chr15_+_101402041 | 0.37 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr4_+_76649797 | 0.37 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr10_+_180643 | 0.36 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr5_-_152069089 | 0.36 |
ENST00000506723.2
|
AC091969.1
|
AC091969.1 |
| chr3_+_107364769 | 0.35 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr4_+_79567057 | 0.34 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr21_+_18811205 | 0.34 |
ENST00000440664.1
|
C21orf37
|
chromosome 21 open reading frame 37 |
| chrX_+_149867681 | 0.34 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr4_-_190948409 | 0.34 |
ENST00000504750.1
ENST00000378763.1 |
FRG2
|
FSHD region gene 2 |
| chr2_+_149632783 | 0.33 |
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C |
| chr11_-_11374904 | 0.32 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr3_+_120461484 | 0.32 |
ENST00000484715.1
ENST00000469772.1 ENST00000283875.5 ENST00000492959.1 |
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa |
| chr11_+_13690249 | 0.32 |
ENST00000532701.1
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr12_-_118628350 | 0.32 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr5_-_31532238 | 0.32 |
ENST00000507438.1
|
DROSHA
|
drosha, ribonuclease type III |
| chr8_+_19796381 | 0.32 |
ENST00000524029.1
ENST00000522701.1 ENST00000311322.8 |
LPL
|
lipoprotein lipase |
| chr19_-_54663473 | 0.32 |
ENST00000222224.3
|
LENG1
|
leukocyte receptor cluster (LRC) member 1 |
| chr1_+_170501270 | 0.31 |
ENST00000367763.3
ENST00000367762.1 |
GORAB
|
golgin, RAB6-interacting |
| chr21_+_17566643 | 0.31 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr5_+_68513622 | 0.31 |
ENST00000512880.1
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chrX_-_55187588 | 0.31 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr12_+_32115400 | 0.31 |
ENST00000381054.3
|
KIAA1551
|
KIAA1551 |
| chr16_-_15149917 | 0.30 |
ENST00000287706.3
|
NTAN1
|
N-terminal asparagine amidase |
| chr17_-_79849438 | 0.30 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr19_+_52932435 | 0.30 |
ENST00000301085.4
|
ZNF534
|
zinc finger protein 534 |
| chr18_-_2982869 | 0.30 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr8_+_86747543 | 0.29 |
ENST00000425429.2
|
REXO1L11P
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 11, pseudogene |
| chr17_+_66511540 | 0.29 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr9_-_6015607 | 0.29 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr2_-_24299308 | 0.28 |
ENST00000233468.4
|
SF3B14
|
Pre-mRNA branch site protein p14 |
| chr2_+_208527094 | 0.28 |
ENST00000429730.1
|
AC079767.4
|
AC079767.4 |
| chr6_-_10838736 | 0.28 |
ENST00000536370.1
ENST00000474039.1 |
MAK
|
male germ cell-associated kinase |
| chr12_-_54653313 | 0.28 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr5_-_74326724 | 0.28 |
ENST00000322348.4
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4, core 2 |
| chr2_+_101437487 | 0.28 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr13_+_21141270 | 0.27 |
ENST00000319980.6
ENST00000537103.1 ENST00000389373.3 |
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr10_-_33281363 | 0.27 |
ENST00000534049.1
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr2_+_29001711 | 0.27 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr13_+_21141208 | 0.27 |
ENST00000351808.5
|
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr5_+_154238149 | 0.27 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr5_+_67535647 | 0.27 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_-_220219775 | 0.27 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr11_-_95522907 | 0.27 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr9_-_75695323 | 0.26 |
ENST00000419959.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr1_-_115124257 | 0.26 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr6_-_75953484 | 0.26 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr12_+_111284764 | 0.26 |
ENST00000545036.1
ENST00000308208.5 |
CCDC63
|
coiled-coil domain containing 63 |
| chr6_-_26199499 | 0.26 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr4_+_148653206 | 0.26 |
ENST00000336498.3
|
ARHGAP10
|
Rho GTPase activating protein 10 |
| chr5_+_142149932 | 0.25 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chrX_+_154444643 | 0.25 |
ENST00000286428.5
|
VBP1
|
von Hippel-Lindau binding protein 1 |
| chr3_+_107364683 | 0.25 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr15_-_75918787 | 0.25 |
ENST00000564086.1
|
SNUPN
|
snurportin 1 |
| chr4_-_14889791 | 0.25 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr1_-_45272951 | 0.25 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr4_-_110651143 | 0.24 |
ENST00000243501.5
|
PLA2G12A
|
phospholipase A2, group XIIA |
| chr11_+_114270752 | 0.24 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr5_+_140079919 | 0.24 |
ENST00000274712.3
|
ZMAT2
|
zinc finger, matrin-type 2 |
| chr9_+_88556444 | 0.24 |
ENST00000376040.1
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr12_+_25348186 | 0.24 |
ENST00000555711.1
ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5
|
LYR motif containing 5 |
| chr11_-_47736896 | 0.23 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr12_+_25348139 | 0.23 |
ENST00000557540.2
ENST00000381356.4 |
LYRM5
|
LYR motif containing 5 |
| chr18_-_74839891 | 0.23 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr5_+_68513557 | 0.23 |
ENST00000256441.4
|
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr2_+_109223595 | 0.23 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr5_-_89705537 | 0.23 |
ENST00000522864.1
ENST00000522083.1 ENST00000522565.1 ENST00000522842.1 ENST00000283122.3 |
CETN3
|
centrin, EF-hand protein, 3 |
| chr8_+_11225911 | 0.23 |
ENST00000284481.3
|
C8orf12
|
chromosome 8 open reading frame 12 |
| chr3_-_39321512 | 0.22 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr19_+_32836499 | 0.22 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr6_+_31939608 | 0.22 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr15_+_58724184 | 0.22 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr10_-_29811456 | 0.22 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr7_-_99277610 | 0.22 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr2_-_201753717 | 0.22 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr20_-_44298878 | 0.22 |
ENST00000324384.3
ENST00000356562.2 |
WFDC11
|
WAP four-disulfide core domain 11 |
| chr2_-_198062758 | 0.22 |
ENST00000328737.2
|
ANKRD44
|
ankyrin repeat domain 44 |
| chr1_+_219347203 | 0.21 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr1_-_198906528 | 0.21 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr6_+_97010424 | 0.21 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chrX_-_115085422 | 0.21 |
ENST00000430756.2
|
RP11-761E20.1
|
RP11-761E20.1 |
| chr15_-_80263506 | 0.21 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr3_-_176914191 | 0.21 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr11_-_89653576 | 0.21 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr4_-_76649546 | 0.21 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr5_+_145826867 | 0.21 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr5_+_32585605 | 0.20 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr8_-_16859690 | 0.20 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr1_-_91487770 | 0.20 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644 |
| chr3_+_190333097 | 0.20 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr16_-_57219966 | 0.20 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr8_+_91233744 | 0.20 |
ENST00000524361.1
|
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr20_-_17539456 | 0.20 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr11_+_121447469 | 0.19 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr1_-_42801540 | 0.19 |
ENST00000372573.1
|
FOXJ3
|
forkhead box J3 |
| chr16_+_30007524 | 0.19 |
ENST00000567254.1
ENST00000567705.1 |
INO80E
|
INO80 complex subunit E |
| chr6_+_3259148 | 0.19 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr2_+_60983361 | 0.19 |
ENST00000238714.3
|
PAPOLG
|
poly(A) polymerase gamma |
| chr1_-_91487806 | 0.19 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr19_-_6393216 | 0.19 |
ENST00000595047.1
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa |
| chr16_+_57220193 | 0.18 |
ENST00000564435.1
ENST00000562959.1 ENST00000394420.4 ENST00000568505.2 ENST00000537866.1 |
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr12_-_110434021 | 0.18 |
ENST00000355312.3
ENST00000551209.1 ENST00000550186.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr5_-_31532160 | 0.18 |
ENST00000511367.2
ENST00000513349.1 |
DROSHA
|
drosha, ribonuclease type III |
| chr5_+_142149955 | 0.18 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr16_+_24549014 | 0.18 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr6_-_10838710 | 0.18 |
ENST00000313243.2
|
MAK
|
male germ cell-associated kinase |
| chr12_-_68845165 | 0.18 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr14_-_102552659 | 0.18 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr4_-_77069573 | 0.18 |
ENST00000264883.3
|
NUP54
|
nucleoporin 54kDa |
| chrX_-_48980098 | 0.17 |
ENST00000156109.5
|
GPKOW
|
G patch domain and KOW motifs |
| chr2_+_201994042 | 0.17 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr11_+_95523621 | 0.17 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr3_+_157827841 | 0.17 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr12_+_122688090 | 0.17 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr15_-_66679019 | 0.17 |
ENST00000568216.1
ENST00000562124.1 ENST00000570251.1 |
TIPIN
|
TIMELESS interacting protein |
| chr11_+_13690200 | 0.17 |
ENST00000354817.3
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr4_-_170924888 | 0.17 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr3_+_118892362 | 0.16 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr16_+_31724552 | 0.16 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr3_-_138725110 | 0.16 |
ENST00000383163.2
|
PRR23A
|
proline rich 23A |
| chr16_-_21875424 | 0.16 |
ENST00000541674.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr14_-_75536182 | 0.16 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr5_+_154238042 | 0.16 |
ENST00000519211.1
ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr12_-_118796971 | 0.16 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr6_+_74104566 | 0.16 |
ENST00000539829.1
|
DDX43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr1_-_154941350 | 0.16 |
ENST00000444179.1
ENST00000414115.1 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr13_+_20207782 | 0.16 |
ENST00000414242.2
ENST00000361479.5 |
MPHOSPH8
|
M-phase phosphoprotein 8 |
| chr9_-_13279563 | 0.16 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr2_+_197577841 | 0.16 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr14_+_55595762 | 0.16 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr1_+_209929494 | 0.16 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr15_-_38519066 | 0.16 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr17_+_61151306 | 0.16 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr12_+_27619743 | 0.16 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr8_+_82192501 | 0.16 |
ENST00000297258.6
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated) |
| chr4_+_40751914 | 0.15 |
ENST00000381782.2
ENST00000316607.5 |
NSUN7
|
NOP2/Sun domain family, member 7 |
| chrX_+_77166172 | 0.15 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr1_+_104159999 | 0.15 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr3_+_107318157 | 0.15 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr12_-_110434183 | 0.15 |
ENST00000360185.4
ENST00000354574.4 ENST00000338373.5 ENST00000343646.5 ENST00000356259.4 ENST00000553118.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chrX_-_148571884 | 0.14 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr10_+_47894572 | 0.14 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr3_+_179280668 | 0.14 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr4_-_74088800 | 0.14 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr15_+_44719970 | 0.14 |
ENST00000558966.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr6_+_150920999 | 0.14 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr15_+_98503922 | 0.14 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr7_+_77325738 | 0.14 |
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like |
| chr12_+_57157100 | 0.14 |
ENST00000322165.1
|
HSD17B6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr5_+_154238096 | 0.14 |
ENST00000517568.1
ENST00000524105.1 ENST00000285896.6 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr18_+_19192228 | 0.14 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr13_-_81801115 | 0.14 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr1_+_92632542 | 0.14 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr12_+_26164645 | 0.14 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr14_-_61447752 | 0.13 |
ENST00000555420.1
ENST00000553903.1 |
TRMT5
|
tRNA methyltransferase 5 |
| chr8_+_91233711 | 0.13 |
ENST00000523283.1
ENST00000517400.1 |
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr11_+_43333513 | 0.13 |
ENST00000534695.1
ENST00000455725.2 ENST00000531273.1 ENST00000420461.2 ENST00000378852.3 ENST00000534600.1 |
API5
|
apoptosis inhibitor 5 |
| chr4_-_6557336 | 0.13 |
ENST00000507294.1
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr12_-_114211474 | 0.13 |
ENST00000550905.1
ENST00000547963.1 |
RP11-438N16.1
|
RP11-438N16.1 |
| chr8_-_62602327 | 0.13 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr5_-_1882858 | 0.13 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr13_-_31191642 | 0.13 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr5_+_31532373 | 0.13 |
ENST00000325366.9
ENST00000355907.3 ENST00000507818.2 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr10_-_48806939 | 0.13 |
ENST00000374233.3
ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B
|
protein tyrosine phosphatase, non-receptor type 20B |
| chr7_+_8008418 | 0.13 |
ENST00000223145.5
|
GLCCI1
|
glucocorticoid induced transcript 1 |
| chr9_-_19065082 | 0.13 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin-like complex, subunit 6 |
| chr6_-_76072719 | 0.13 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr12_+_8662057 | 0.13 |
ENST00000382064.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr12_-_110434096 | 0.13 |
ENST00000320063.9
ENST00000457474.2 ENST00000547815.1 ENST00000361006.5 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DUXA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.8 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.2 | 0.7 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.4 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.3 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.5 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.5 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.1 | 0.3 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.1 | 0.2 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.2 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.3 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.3 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.2 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) cellular response to lead ion(GO:0071284) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.5 | GO:2000628 | production of siRNA involved in RNA interference(GO:0030422) regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.2 | GO:2000521 | positive regulation of mononuclear cell migration(GO:0071677) negative regulation of immunological synapse formation(GO:2000521) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0060979 | transforming growth factor beta receptor complex assembly(GO:0007181) vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.1 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.0 | 0.8 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.5 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.3 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.2 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0031947 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 1.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.4 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.2 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.4 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |