Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
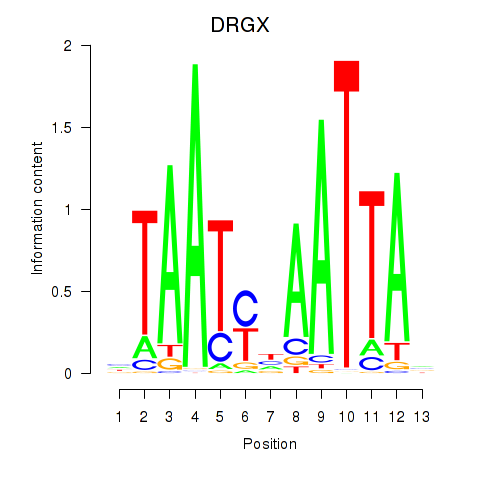
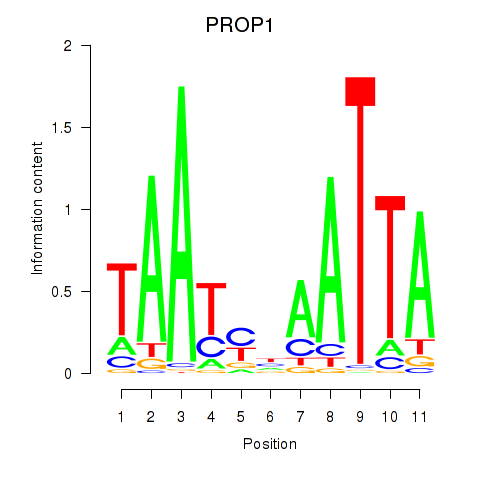
Results for DRGX_PROP1
Z-value: 0.50
Transcription factors associated with DRGX_PROP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DRGX
|
ENSG00000165606.4 | dorsal root ganglia homeobox |
|
PROP1
|
ENSG00000175325.2 | PROP paired-like homeobox 1 |
Activity profile of DRGX_PROP1 motif
Sorted Z-values of DRGX_PROP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_61151306 | 0.67 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr4_+_155484103 | 0.36 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr6_-_107235331 | 0.35 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr3_-_195310802 | 0.32 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr14_-_75083313 | 0.26 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr15_+_101417919 | 0.25 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr3_-_12587055 | 0.24 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr12_-_75784669 | 0.23 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr4_-_89951028 | 0.23 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr14_-_75536182 | 0.22 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr12_-_118796971 | 0.22 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr17_+_71228843 | 0.21 |
ENST00000582391.1
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr4_+_76649753 | 0.20 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr11_-_96076334 | 0.19 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr19_-_54618650 | 0.19 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr6_-_107235378 | 0.19 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr4_+_155484155 | 0.18 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr12_+_14572070 | 0.18 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr3_+_159943362 | 0.17 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr12_-_118797475 | 0.16 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_12007897 | 0.16 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr2_+_191334212 | 0.15 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr14_+_21566980 | 0.15 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr13_-_81801115 | 0.13 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr18_-_2982869 | 0.12 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr10_-_31146615 | 0.12 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438 |
| chr6_-_76072719 | 0.11 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr12_-_91546926 | 0.11 |
ENST00000550758.1
|
DCN
|
decorin |
| chr17_+_74463650 | 0.11 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr3_-_187455680 | 0.11 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr5_-_87516448 | 0.11 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr5_-_111312622 | 0.11 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr3_+_138340067 | 0.10 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr2_-_114461655 | 0.10 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr8_+_67039131 | 0.10 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr16_+_53133070 | 0.10 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_88556444 | 0.10 |
ENST00000376040.1
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr10_-_75226166 | 0.10 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr2_+_102413726 | 0.09 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr3_+_138340049 | 0.09 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_+_109756523 | 0.09 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr10_-_29923893 | 0.09 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr2_+_44502630 | 0.09 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr3_-_87325612 | 0.09 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr17_+_71228537 | 0.09 |
ENST00000577615.1
ENST00000585109.1 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr11_+_108094786 | 0.09 |
ENST00000601453.1
|
ATM
|
ataxia telangiectasia mutated |
| chr4_-_103940791 | 0.08 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr5_+_59783941 | 0.08 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr5_+_140514782 | 0.08 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr7_-_107883678 | 0.08 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr10_+_67330096 | 0.08 |
ENST00000433152.4
ENST00000601979.1 ENST00000599409.1 ENST00000608678.1 |
RP11-222A11.1
|
RP11-222A11.1 |
| chr7_-_117512264 | 0.08 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr11_+_18433840 | 0.08 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr6_+_3259148 | 0.08 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr1_-_151148442 | 0.07 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr11_+_1295809 | 0.07 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr15_+_44092784 | 0.07 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chr6_+_26087646 | 0.07 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chrX_+_100878112 | 0.07 |
ENST00000491568.2
ENST00000479298.1 |
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr12_-_118628350 | 0.07 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr16_+_81272287 | 0.07 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr4_+_142558078 | 0.07 |
ENST00000529613.1
|
IL15
|
interleukin 15 |
| chr6_+_88106840 | 0.06 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr1_+_92632542 | 0.06 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr1_-_242162375 | 0.06 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr12_+_59989918 | 0.06 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr9_+_125795788 | 0.06 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr11_-_111649015 | 0.06 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr7_-_27169801 | 0.06 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr1_+_225600404 | 0.06 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr3_+_111393659 | 0.06 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_+_8040739 | 0.06 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr10_-_25304889 | 0.06 |
ENST00000483339.2
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr2_-_183291741 | 0.06 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr16_+_53469525 | 0.06 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr9_+_2159850 | 0.06 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_111393501 | 0.06 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr2_-_172087824 | 0.06 |
ENST00000521943.1
|
TLK1
|
tousled-like kinase 1 |
| chr1_-_200379129 | 0.06 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr15_+_44719996 | 0.05 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr2_-_17981462 | 0.05 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr5_+_59783540 | 0.05 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr8_-_57123815 | 0.05 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr11_-_111649074 | 0.05 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr12_+_122688090 | 0.05 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr3_-_9291063 | 0.05 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr1_+_62439037 | 0.05 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr6_-_107235287 | 0.05 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr5_+_142149932 | 0.05 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr19_-_58485895 | 0.05 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr2_+_179317994 | 0.05 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr2_+_152214098 | 0.05 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr14_+_75536280 | 0.05 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr7_-_82792215 | 0.05 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr13_-_47471155 | 0.05 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr12_+_100897130 | 0.04 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_+_140079919 | 0.04 |
ENST00000274712.3
|
ZMAT2
|
zinc finger, matrin-type 2 |
| chr3_-_33686925 | 0.04 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_-_76649546 | 0.04 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chrX_+_119737806 | 0.04 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr5_-_59783882 | 0.04 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr10_+_115312766 | 0.04 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr13_-_88463487 | 0.04 |
ENST00000606221.1
|
RP11-471M2.3
|
RP11-471M2.3 |
| chr9_+_124329336 | 0.04 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr17_-_56082455 | 0.04 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr2_+_54683419 | 0.04 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr3_-_156878482 | 0.04 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr3_-_141747950 | 0.04 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr22_-_30953587 | 0.04 |
ENST00000453479.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr12_+_19358228 | 0.04 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr8_+_92114060 | 0.04 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr8_+_26150628 | 0.04 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr19_-_54619006 | 0.04 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr2_+_109237717 | 0.03 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr14_+_101295638 | 0.03 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr9_-_34397800 | 0.03 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr8_-_112248400 | 0.03 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr17_-_71228357 | 0.03 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr11_-_7847519 | 0.03 |
ENST00000328375.1
|
OR5P3
|
olfactory receptor, family 5, subfamily P, member 3 |
| chrX_+_100878079 | 0.03 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr14_+_39944025 | 0.03 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr10_+_106034637 | 0.03 |
ENST00000401888.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr6_-_24358264 | 0.03 |
ENST00000378454.3
|
DCDC2
|
doublecortin domain containing 2 |
| chr17_-_14683517 | 0.03 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr4_+_160188306 | 0.03 |
ENST00000510510.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr8_-_10512569 | 0.03 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr15_+_44719970 | 0.03 |
ENST00000558966.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr15_+_66874502 | 0.03 |
ENST00000558797.1
|
RP11-321F6.1
|
HCG2003567; Uncharacterized protein |
| chr17_-_38956205 | 0.03 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr5_+_98109322 | 0.03 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr7_-_111032971 | 0.03 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr17_-_39623681 | 0.03 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr6_-_137314371 | 0.03 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr2_+_75873902 | 0.03 |
ENST00000393909.2
ENST00000358788.6 ENST00000409374.1 |
MRPL19
|
mitochondrial ribosomal protein L19 |
| chrX_+_10126488 | 0.03 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr9_+_44867571 | 0.03 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr10_+_24544249 | 0.03 |
ENST00000430453.2
|
KIAA1217
|
KIAA1217 |
| chr7_-_80551671 | 0.03 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr1_+_84630574 | 0.03 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr15_+_44719790 | 0.03 |
ENST00000558791.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr20_+_32150140 | 0.03 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr2_+_136343943 | 0.03 |
ENST00000409606.1
|
R3HDM1
|
R3H domain containing 1 |
| chr11_+_57480046 | 0.03 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr9_-_39239171 | 0.03 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr8_-_10987745 | 0.03 |
ENST00000400102.3
|
AF131215.5
|
AF131215.5 |
| chr17_+_44701402 | 0.03 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr1_+_180199393 | 0.02 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr21_-_15583165 | 0.02 |
ENST00000536861.1
|
LIPI
|
lipase, member I |
| chr5_+_142149955 | 0.02 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr14_-_38064198 | 0.02 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr4_-_187647773 | 0.02 |
ENST00000509647.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr4_+_113568207 | 0.02 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr22_-_27456361 | 0.02 |
ENST00000453934.1
|
CTA-992D9.6
|
CTA-992D9.6 |
| chr4_+_76649797 | 0.02 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr17_+_57807062 | 0.02 |
ENST00000587259.1
|
VMP1
|
vacuole membrane protein 1 |
| chr5_-_137475071 | 0.02 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr12_-_57873631 | 0.02 |
ENST00000393791.3
ENST00000356411.2 ENST00000552249.1 |
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr3_+_186692745 | 0.02 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr5_+_140743859 | 0.02 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr15_-_43398274 | 0.02 |
ENST00000382177.2
ENST00000290650.4 |
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chrX_+_77166172 | 0.02 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr6_-_111804905 | 0.02 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr17_+_71228793 | 0.02 |
ENST00000426147.2
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr10_-_128359074 | 0.02 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr5_+_140227357 | 0.02 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr3_-_165555200 | 0.02 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr10_+_53806501 | 0.02 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr1_-_157069590 | 0.02 |
ENST00000454449.2
|
ETV3L
|
ets variant 3-like |
| chr3_-_191000172 | 0.02 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr15_+_57511609 | 0.02 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr3_-_27763803 | 0.02 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr1_+_78383813 | 0.02 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr4_+_160188889 | 0.02 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr15_-_52970820 | 0.02 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr12_-_15815626 | 0.02 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr4_-_143227088 | 0.01 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr9_+_21440440 | 0.01 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr3_-_98241760 | 0.01 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr4_+_74347400 | 0.01 |
ENST00000226355.3
|
AFM
|
afamin |
| chr21_+_17792672 | 0.01 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_-_21875424 | 0.01 |
ENST00000541674.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr3_-_156878540 | 0.01 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr3_-_33686743 | 0.01 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_-_87325728 | 0.01 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr16_-_74700737 | 0.01 |
ENST00000576652.1
ENST00000572337.1 ENST00000571750.1 ENST00000572990.1 ENST00000361070.4 |
RFWD3
|
ring finger and WD repeat domain 3 |
| chr12_+_54410664 | 0.01 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr4_-_143226979 | 0.01 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr8_-_62602327 | 0.01 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr1_+_174933899 | 0.01 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr3_-_151034734 | 0.01 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr12_+_72080253 | 0.01 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr8_+_86999516 | 0.01 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr5_-_111093759 | 0.01 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr4_-_184243561 | 0.01 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr5_-_55412774 | 0.01 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chrX_-_11284095 | 0.01 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr7_+_123241908 | 0.01 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr6_-_41039567 | 0.01 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr12_+_58003935 | 0.01 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr8_+_77318769 | 0.01 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DRGX_PROP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.3 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.0 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.0 | GO:0090294 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |