Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
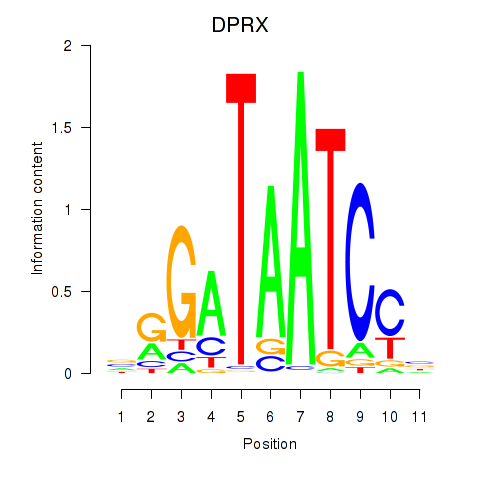
Results for DPRX
Z-value: 0.97
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | divergent-paired related homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DPRX | hg19_v2_chr19_+_54135310_54135310 | 0.13 | 8.7e-01 | Click! |
Activity profile of DPRX motif
Sorted Z-values of DPRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_321340 | 1.91 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr11_-_321050 | 0.96 |
ENST00000399808.4
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr5_+_149865377 | 0.46 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr10_+_104178946 | 0.36 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr7_-_75401513 | 0.34 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr11_-_104916034 | 0.29 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr1_-_12679171 | 0.29 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr15_+_42565393 | 0.28 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr2_+_120517717 | 0.28 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr4_+_147096837 | 0.26 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr17_-_65992544 | 0.25 |
ENST00000580729.1
|
RP11-855A2.5
|
RP11-855A2.5 |
| chr19_-_44123734 | 0.24 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr8_+_94752349 | 0.24 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr19_+_7981030 | 0.22 |
ENST00000565886.1
|
TGFBR3L
|
transforming growth factor, beta receptor III-like |
| chrX_-_77150911 | 0.21 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr1_-_153521597 | 0.21 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr2_+_27799389 | 0.20 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr7_-_155601766 | 0.19 |
ENST00000430104.1
|
SHH
|
sonic hedgehog |
| chr1_+_36024107 | 0.18 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr8_+_145597713 | 0.18 |
ENST00000308860.6
ENST00000532190.1 |
ADCK5
|
aarF domain containing kinase 5 |
| chr19_-_44124019 | 0.18 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chrX_+_100878112 | 0.17 |
ENST00000491568.2
ENST00000479298.1 |
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr7_-_97881429 | 0.17 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr2_+_128175997 | 0.17 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr15_+_29211570 | 0.16 |
ENST00000558804.1
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr1_-_762885 | 0.16 |
ENST00000536430.1
ENST00000473798.1 |
LINC00115
|
long intergenic non-protein coding RNA 115 |
| chr17_+_48351785 | 0.16 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr2_-_176046391 | 0.16 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr15_+_42565464 | 0.16 |
ENST00000562170.1
ENST00000562859.1 |
GANC
|
glucosidase, alpha; neutral C |
| chr1_-_153521714 | 0.16 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr1_-_219615984 | 0.16 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr4_+_159131630 | 0.15 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr1_+_200863949 | 0.15 |
ENST00000413687.2
|
C1orf106
|
chromosome 1 open reading frame 106 |
| chr3_-_54962100 | 0.15 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr7_+_7222157 | 0.15 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr5_+_118604439 | 0.15 |
ENST00000388882.5
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr1_+_36023370 | 0.15 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr16_+_81272287 | 0.14 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr1_-_150208320 | 0.14 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_+_55183261 | 0.14 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr1_+_164600184 | 0.14 |
ENST00000482110.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr5_+_118604385 | 0.14 |
ENST00000274456.6
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr12_+_113416265 | 0.13 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr14_-_22005197 | 0.13 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr12_-_111395610 | 0.13 |
ENST00000548329.1
ENST00000546852.1 |
RP1-46F2.3
|
RP1-46F2.3 |
| chr14_-_94789663 | 0.13 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr17_-_47925379 | 0.12 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr20_-_36794938 | 0.12 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr19_+_36139125 | 0.12 |
ENST00000246554.3
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr9_+_131133598 | 0.12 |
ENST00000372853.4
ENST00000452446.1 ENST00000372850.1 ENST00000372847.1 |
URM1
|
ubiquitin related modifier 1 |
| chrX_+_15767971 | 0.12 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr11_+_64949343 | 0.11 |
ENST00000279247.6
ENST00000532285.1 ENST00000534373.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr4_+_159131596 | 0.11 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr17_+_8191815 | 0.11 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr19_-_50370799 | 0.11 |
ENST00000600910.1
ENST00000322344.3 ENST00000600573.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr19_-_4831701 | 0.11 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr20_+_33759854 | 0.11 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chr11_+_61248583 | 0.11 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr9_+_4839762 | 0.10 |
ENST00000448872.2
ENST00000441844.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr1_-_161014731 | 0.10 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr2_-_216003127 | 0.10 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr19_+_56165480 | 0.09 |
ENST00000450554.2
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr7_-_135412925 | 0.09 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr5_-_77072145 | 0.09 |
ENST00000380377.4
|
TBCA
|
tubulin folding cofactor A |
| chr15_+_59908633 | 0.09 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr16_-_15950868 | 0.09 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr15_+_22368478 | 0.09 |
ENST00000332663.2
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2 |
| chr11_+_61522844 | 0.09 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr4_-_103998060 | 0.09 |
ENST00000339611.4
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr12_+_113344755 | 0.09 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr7_+_30323923 | 0.08 |
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2 |
| chr2_+_33701286 | 0.08 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_+_19638788 | 0.08 |
ENST00000375155.3
ENST00000375153.3 ENST00000400548.2 |
PQLC2
|
PQ loop repeat containing 2 |
| chr3_+_49058444 | 0.08 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr17_-_4643161 | 0.08 |
ENST00000574412.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr2_+_74685413 | 0.08 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr12_+_133563026 | 0.08 |
ENST00000540238.1
ENST00000328654.5 ENST00000544181.1 |
ZNF26
|
zinc finger protein 26 |
| chr6_-_32908792 | 0.08 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr1_+_210501589 | 0.08 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr2_+_145425534 | 0.08 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr22_-_36903069 | 0.08 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr19_-_42758040 | 0.08 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr15_-_83240783 | 0.08 |
ENST00000568994.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr20_-_36794902 | 0.08 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr11_+_65343494 | 0.08 |
ENST00000309295.4
ENST00000533237.1 |
EHBP1L1
|
EH domain binding protein 1-like 1 |
| chr8_+_72755367 | 0.08 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr11_-_104972158 | 0.08 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr1_-_98515395 | 0.07 |
ENST00000424528.2
|
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr2_-_98280383 | 0.07 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr8_+_86999516 | 0.07 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr8_+_38244638 | 0.07 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr2_+_69201705 | 0.07 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr11_+_60681346 | 0.07 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr19_+_36132631 | 0.07 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr14_+_50234309 | 0.07 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr9_-_95244781 | 0.07 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr3_+_141105705 | 0.07 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr19_+_41284121 | 0.07 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr11_+_111385497 | 0.07 |
ENST00000375618.4
ENST00000529167.1 ENST00000332814.6 |
C11orf88
|
chromosome 11 open reading frame 88 |
| chr7_-_99869799 | 0.07 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr11_+_64949158 | 0.06 |
ENST00000527739.1
ENST00000526966.1 ENST00000533129.1 ENST00000524773.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr8_+_48173556 | 0.06 |
ENST00000524006.1
|
SPIDR
|
scaffolding protein involved in DNA repair |
| chr6_+_26383404 | 0.06 |
ENST00000416795.2
ENST00000494184.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr16_-_4665023 | 0.06 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr9_-_35096545 | 0.06 |
ENST00000378617.3
ENST00000341666.3 ENST00000361778.2 |
PIGO
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr2_-_190044480 | 0.06 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr15_+_42565844 | 0.06 |
ENST00000566442.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr1_+_205538165 | 0.06 |
ENST00000536357.1
|
MFSD4
|
major facilitator superfamily domain containing 4 |
| chr2_+_145425573 | 0.06 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr4_-_164534657 | 0.06 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr1_-_170043709 | 0.06 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr7_-_84121858 | 0.06 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr19_-_18709357 | 0.06 |
ENST00000597131.1
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr16_-_4664860 | 0.06 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr4_-_170924888 | 0.06 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr12_+_122516626 | 0.06 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr3_-_15469045 | 0.06 |
ENST00000450816.2
|
METTL6
|
methyltransferase like 6 |
| chr1_+_81771806 | 0.05 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr8_+_145215928 | 0.05 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr5_-_54988448 | 0.05 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr11_-_61849656 | 0.05 |
ENST00000524942.1
|
RP11-810P12.5
|
RP11-810P12.5 |
| chr1_+_150039877 | 0.05 |
ENST00000419023.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr18_-_32957260 | 0.05 |
ENST00000587422.1
ENST00000306346.1 ENST00000589332.1 ENST00000586687.1 ENST00000585522.1 |
ZNF396
|
zinc finger protein 396 |
| chrX_+_99899180 | 0.05 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr19_+_55738587 | 0.05 |
ENST00000598855.1
|
AC010327.2
|
Uncharacterized protein; cDNA FLJ45856 fis, clone OCBBF2025631 |
| chr10_-_105845674 | 0.05 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr7_+_28452130 | 0.05 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr1_-_22263790 | 0.05 |
ENST00000374695.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr18_-_52989525 | 0.05 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr17_+_41150290 | 0.05 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr18_-_52989217 | 0.05 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr11_-_85430163 | 0.05 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr16_-_21416640 | 0.05 |
ENST00000542817.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr1_+_207262170 | 0.05 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr5_+_139781393 | 0.04 |
ENST00000360839.2
ENST00000297183.6 ENST00000421134.1 ENST00000394723.3 ENST00000511151.1 |
ANKHD1
|
ankyrin repeat and KH domain containing 1 |
| chr2_+_33661382 | 0.04 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_+_16767195 | 0.04 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr4_+_169418255 | 0.04 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_+_77021702 | 0.04 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr1_-_158656488 | 0.04 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr19_+_10531150 | 0.04 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr5_+_35856951 | 0.04 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr3_-_112564797 | 0.04 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr11_+_64948665 | 0.04 |
ENST00000533820.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr12_-_49523896 | 0.04 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr4_+_159131346 | 0.04 |
ENST00000508243.1
ENST00000296529.6 |
TMEM144
|
transmembrane protein 144 |
| chr11_-_104905840 | 0.04 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr16_-_21849091 | 0.04 |
ENST00000537951.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chrX_-_102757802 | 0.04 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr12_-_124419531 | 0.04 |
ENST00000514254.2
|
DNAH10OS
|
dynein, axonemal, heavy chain 10 opposite strand |
| chr14_-_101034407 | 0.04 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr3_-_178976996 | 0.04 |
ENST00000485523.1
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr3_+_188664988 | 0.04 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr12_-_113574028 | 0.04 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr17_+_7482785 | 0.04 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr11_-_8816375 | 0.04 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr1_-_110950255 | 0.04 |
ENST00000483260.1
ENST00000474861.2 ENST00000602318.1 |
LAMTOR5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr2_-_85555385 | 0.04 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr6_+_42018614 | 0.04 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr17_-_37557846 | 0.04 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr8_-_110656995 | 0.04 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr12_+_6949964 | 0.04 |
ENST00000541978.1
ENST00000435982.2 |
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr1_-_27481401 | 0.04 |
ENST00000263980.3
|
SLC9A1
|
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
| chr2_+_26915584 | 0.04 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr11_+_61560348 | 0.04 |
ENST00000535723.1
ENST00000574708.1 |
FEN1
FADS2
|
flap structure-specific endonuclease 1 fatty acid desaturase 2 |
| chr4_+_170541835 | 0.03 |
ENST00000504131.2
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr5_+_140345820 | 0.03 |
ENST00000289269.5
|
PCDHAC2
|
protocadherin alpha subfamily C, 2 |
| chr7_+_55980331 | 0.03 |
ENST00000429591.2
|
ZNF713
|
zinc finger protein 713 |
| chr10_-_105845536 | 0.03 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr5_+_139781445 | 0.03 |
ENST00000532219.1
ENST00000394722.3 |
ANKHD1-EIF4EBP3
ANKHD1
|
ANKHD1-EIF4EBP3 readthrough ankyrin repeat and KH domain containing 1 |
| chr17_-_19771242 | 0.03 |
ENST00000361658.2
|
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr12_+_124997766 | 0.03 |
ENST00000543970.1
|
RP11-83B20.1
|
RP11-83B20.1 |
| chr10_-_116418053 | 0.03 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr16_+_21244986 | 0.03 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr6_+_26383318 | 0.03 |
ENST00000469230.1
ENST00000490025.1 ENST00000356709.4 ENST00000352867.2 ENST00000493275.1 ENST00000472507.1 ENST00000482536.1 ENST00000432533.2 ENST00000482842.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr6_+_36164487 | 0.03 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr4_-_103998439 | 0.03 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr17_-_1928621 | 0.03 |
ENST00000331238.6
|
RTN4RL1
|
reticulon 4 receptor-like 1 |
| chr17_+_5323492 | 0.03 |
ENST00000405578.4
ENST00000574003.1 |
RPAIN
|
RPA interacting protein |
| chr1_+_74701062 | 0.03 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr4_-_103997862 | 0.03 |
ENST00000394785.3
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr19_+_21106081 | 0.03 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr14_+_85996471 | 0.03 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr17_+_7531281 | 0.03 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr1_-_13452656 | 0.03 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr3_+_111393501 | 0.03 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr9_-_5830768 | 0.03 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr1_-_26633480 | 0.03 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr14_+_85996507 | 0.03 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_+_7107999 | 0.03 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr2_-_119605253 | 0.03 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr1_-_46089639 | 0.02 |
ENST00000445048.2
|
CCDC17
|
coiled-coil domain containing 17 |
| chr9_+_116207007 | 0.02 |
ENST00000374140.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr1_-_68516393 | 0.02 |
ENST00000395201.1
|
DIRAS3
|
DIRAS family, GTP-binding RAS-like 3 |
| chr4_-_147442817 | 0.02 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr9_-_23821273 | 0.02 |
ENST00000380110.4
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr17_+_4643300 | 0.02 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chrX_-_49121165 | 0.02 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr12_+_104359641 | 0.02 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr14_-_22005343 | 0.02 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DPRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.2 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.5 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.4 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.4 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.1 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 3.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.5 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.5 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.0 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |