Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
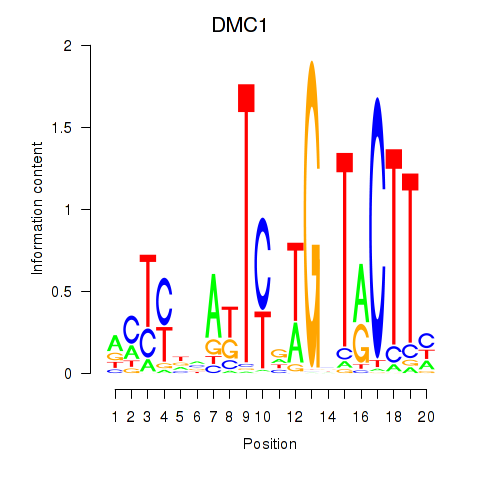
Results for DMC1
Z-value: 0.49
Transcription factors associated with DMC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMC1
|
ENSG00000100206.5 | DNA meiotic recombinase 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMC1 | hg19_v2_chr22_-_38966123_38966163 | -0.61 | 3.9e-01 | Click! |
Activity profile of DMC1 motif
Sorted Z-values of DMC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_42875115 | 0.21 |
ENST00000591137.1
|
CTC-296K1.4
|
CTC-296K1.4 |
| chr12_-_120805872 | 0.18 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr17_+_57297807 | 0.16 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr21_-_35281399 | 0.15 |
ENST00000418933.1
|
ATP5O
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr5_-_66942617 | 0.14 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr2_-_31637560 | 0.13 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr11_-_73720276 | 0.13 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr3_-_156840776 | 0.12 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr12_-_109025849 | 0.11 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr3_+_9944303 | 0.10 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr3_+_9944492 | 0.10 |
ENST00000383814.3
ENST00000454190.2 ENST00000454992.1 |
IL17RE
|
interleukin 17 receptor E |
| chr11_-_73720122 | 0.09 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr17_+_77018896 | 0.08 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_-_3600549 | 0.08 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr2_-_197675000 | 0.07 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr8_+_67104940 | 0.07 |
ENST00000517689.1
|
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr11_-_85779971 | 0.06 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_+_187454749 | 0.06 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr21_-_46348626 | 0.06 |
ENST00000517563.1
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr6_+_88054530 | 0.06 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr10_+_74870206 | 0.06 |
ENST00000357321.4
ENST00000349051.5 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr17_+_38333263 | 0.06 |
ENST00000456989.2
ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr11_-_85780086 | 0.05 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_-_21492113 | 0.05 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr2_+_219246746 | 0.05 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr1_-_19811132 | 0.05 |
ENST00000433834.1
|
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr21_-_46348694 | 0.05 |
ENST00000355153.4
ENST00000397850.2 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr2_+_48667983 | 0.05 |
ENST00000449090.2
|
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr15_-_49103235 | 0.05 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr10_+_62538248 | 0.05 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr2_+_61244697 | 0.04 |
ENST00000401576.1
ENST00000295030.5 ENST00000414712.2 |
PEX13
|
peroxisomal biogenesis factor 13 |
| chr14_-_21491477 | 0.04 |
ENST00000298684.5
ENST00000557169.1 ENST00000553563.1 |
NDRG2
|
NDRG family member 2 |
| chr10_+_69865866 | 0.04 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr7_-_15601595 | 0.04 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr8_+_145438870 | 0.04 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr10_-_135187193 | 0.04 |
ENST00000368547.3
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr2_+_136343904 | 0.04 |
ENST00000436436.1
|
R3HDM1
|
R3H domain containing 1 |
| chr2_+_233497931 | 0.03 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr20_+_2796948 | 0.03 |
ENST00000361033.1
ENST00000380585.1 |
TMEM239
|
transmembrane protein 239 |
| chr10_+_74870253 | 0.03 |
ENST00000544879.1
ENST00000537969.1 ENST00000372997.3 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr2_+_48667898 | 0.03 |
ENST00000281394.4
ENST00000294952.8 |
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr16_+_2255710 | 0.03 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr4_-_76944621 | 0.03 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr11_+_101785727 | 0.03 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr6_-_31670723 | 0.02 |
ENST00000440843.2
ENST00000375842.4 |
ABHD16A
|
abhydrolase domain containing 16A |
| chr10_+_17851362 | 0.02 |
ENST00000331429.2
ENST00000457317.1 |
MRC1L1
|
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
| chr2_-_21022818 | 0.02 |
ENST00000440866.2
ENST00000541941.1 ENST00000402479.2 ENST00000435420.2 ENST00000432947.1 ENST00000403006.2 ENST00000419825.2 ENST00000381090.3 ENST00000237822.3 ENST00000412261.1 |
C2orf43
|
chromosome 2 open reading frame 43 |
| chr6_-_150067696 | 0.02 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr3_-_99569821 | 0.02 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr12_-_71031185 | 0.01 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr1_+_12290121 | 0.01 |
ENST00000358136.3
ENST00000356315.4 |
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr12_-_8803128 | 0.01 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr14_-_68157084 | 0.01 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
| chr22_+_21133469 | 0.01 |
ENST00000406799.1
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr16_-_85969774 | 0.01 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr10_-_74856608 | 0.01 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr14_+_74318513 | 0.01 |
ENST00000555228.1
ENST00000555661.1 |
PTGR2
|
prostaglandin reductase 2 |
| chr14_+_74318611 | 0.01 |
ENST00000555976.1
ENST00000267568.4 |
PTGR2
|
prostaglandin reductase 2 |
| chr17_-_60142609 | 0.01 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr7_+_107332334 | 0.01 |
ENST00000541474.1
ENST00000544569.1 |
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr12_-_71031220 | 0.01 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr14_-_21492251 | 0.01 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr10_+_11047259 | 0.01 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr7_+_6793740 | 0.01 |
ENST00000403107.1
ENST00000404077.1 ENST00000435395.1 ENST00000418406.1 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr1_-_47131521 | 0.01 |
ENST00000542495.1
ENST00000532925.1 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr12_-_50677255 | 0.01 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr2_+_201173667 | 0.01 |
ENST00000409755.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_-_75788038 | 0.00 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr2_-_85641162 | 0.00 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr2_+_171640291 | 0.00 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr7_+_142919130 | 0.00 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr17_+_74075263 | 0.00 |
ENST00000334586.5
ENST00000392503.2 |
ZACN
|
zinc activated ligand-gated ion channel |
| chr6_-_150067632 | 0.00 |
ENST00000460354.2
ENST00000367404.4 ENST00000543637.1 |
NUP43
|
nucleoporin 43kDa |
| chr7_-_6010263 | 0.00 |
ENST00000455618.2
ENST00000405415.1 ENST00000404406.1 ENST00000542644.1 |
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr8_-_101734907 | 0.00 |
ENST00000318607.5
ENST00000521865.1 ENST00000520804.1 ENST00000522720.1 ENST00000521067.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
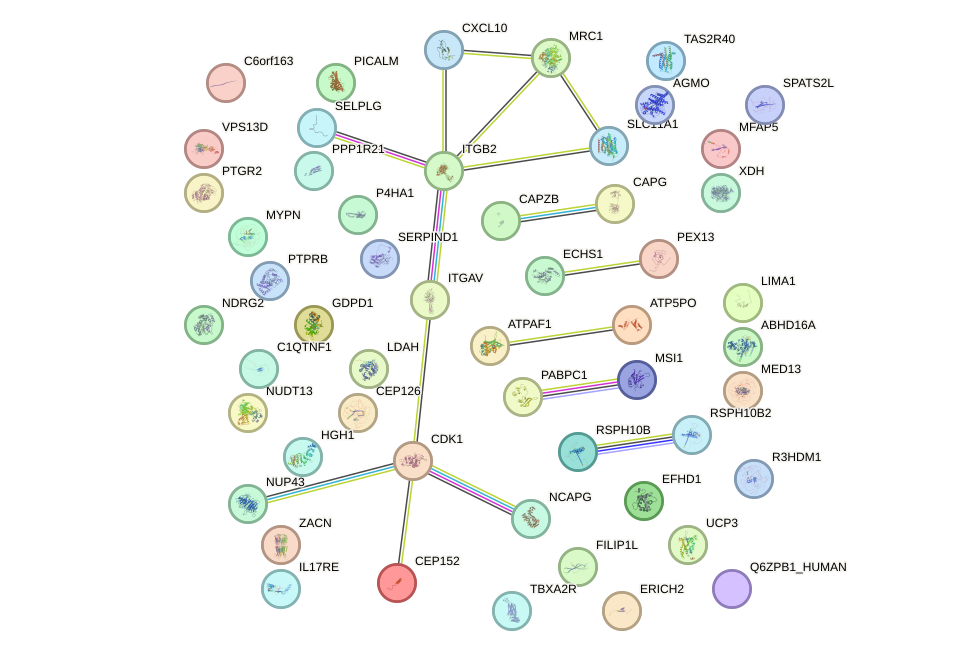
Network of associatons between targets according to the STRING database.
First level regulatory network of DMC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.2 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.0 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |