Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
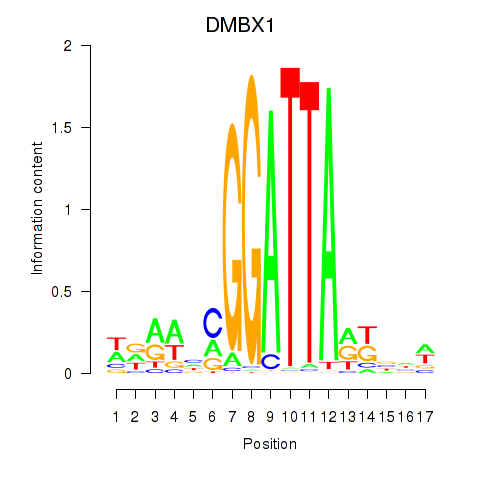
Results for DMBX1
Z-value: 0.43
Transcription factors associated with DMBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMBX1
|
ENSG00000197587.6 | diencephalon/mesencephalon homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMBX1 | hg19_v2_chr1_+_46972668_46972669 | 0.87 | 1.3e-01 | Click! |
Activity profile of DMBX1 motif
Sorted Z-values of DMBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_137851220 | 0.29 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr17_-_19651598 | 0.28 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr3_+_188664988 | 0.27 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr19_+_41284121 | 0.20 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr6_-_26250835 | 0.19 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr3_+_49058444 | 0.17 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_+_46001697 | 0.15 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr1_+_81001398 | 0.14 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr1_-_16563641 | 0.14 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1 |
| chr3_+_111393501 | 0.14 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr2_-_70780572 | 0.13 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr6_-_33297013 | 0.13 |
ENST00000453407.1
|
DAXX
|
death-domain associated protein |
| chr11_-_45940343 | 0.12 |
ENST00000532681.1
|
PEX16
|
peroxisomal biogenesis factor 16 |
| chr2_-_27486951 | 0.12 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr8_-_23282797 | 0.10 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr17_-_19651654 | 0.10 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr2_-_70780770 | 0.10 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr19_-_6459746 | 0.09 |
ENST00000301454.4
ENST00000334510.5 |
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr1_+_180875711 | 0.09 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr8_-_23282820 | 0.09 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr2_-_220252068 | 0.09 |
ENST00000430206.1
ENST00000429013.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr14_+_50234309 | 0.08 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr16_-_31076273 | 0.08 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr5_-_54988448 | 0.07 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chrX_-_102565932 | 0.07 |
ENST00000372674.1
ENST00000372677.3 |
BEX2
|
brain expressed X-linked 2 |
| chr16_-_31076332 | 0.07 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr3_-_149293990 | 0.07 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr17_-_19651668 | 0.07 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr19_+_17448348 | 0.06 |
ENST00000324894.8
ENST00000358792.7 ENST00000600625.1 |
GTPBP3
|
GTP binding protein 3 (mitochondrial) |
| chr3_+_49057876 | 0.06 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chrX_-_48931648 | 0.06 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr17_-_4643161 | 0.06 |
ENST00000574412.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr12_-_56321649 | 0.05 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr9_+_74920335 | 0.05 |
ENST00000451596.2
ENST00000436054.1 |
RP11-63P12.6
|
RP11-63P12.6 |
| chr1_-_47655686 | 0.05 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr1_-_28384598 | 0.05 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr15_-_85201779 | 0.05 |
ENST00000360476.3
ENST00000394588.3 |
NMB
|
neuromedin B |
| chr15_-_61521495 | 0.05 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr12_-_56321397 | 0.05 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr22_+_31002990 | 0.05 |
ENST00000423350.1
|
TCN2
|
transcobalamin II |
| chr17_-_48207157 | 0.04 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr12_-_118628315 | 0.04 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr3_-_49058479 | 0.04 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chrX_-_64754611 | 0.03 |
ENST00000374807.5
ENST00000374811.3 ENST00000374804.5 ENST00000312391.8 |
LAS1L
|
LAS1-like (S. cerevisiae) |
| chr8_-_110656995 | 0.03 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr1_+_162351503 | 0.03 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr15_-_60695071 | 0.03 |
ENST00000557904.1
|
ANXA2
|
annexin A2 |
| chr18_+_29027696 | 0.03 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr5_+_50678921 | 0.03 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr3_+_111393659 | 0.03 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_+_61248583 | 0.03 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr2_-_220252530 | 0.03 |
ENST00000521459.1
|
DNPEP
|
aspartyl aminopeptidase |
| chr16_-_20338748 | 0.03 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr12_-_56727487 | 0.02 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr7_-_99774945 | 0.02 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr12_+_53693812 | 0.02 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr12_-_21487829 | 0.02 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr4_+_140586922 | 0.02 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr10_-_101491828 | 0.02 |
ENST00000370483.5
ENST00000016171.5 |
COX15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr5_-_54988559 | 0.01 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr1_-_13452656 | 0.01 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr3_-_10362725 | 0.01 |
ENST00000397109.3
ENST00000428626.1 ENST00000445064.1 ENST00000431352.1 ENST00000397117.1 ENST00000337354.4 ENST00000383801.2 ENST00000432213.1 ENST00000350697.3 |
SEC13
|
SEC13 homolog (S. cerevisiae) |
| chr9_+_74920408 | 0.01 |
ENST00000451152.1
|
RP11-63P12.6
|
RP11-63P12.6 |
| chr1_-_45965525 | 0.01 |
ENST00000488405.2
ENST00000490551.3 ENST00000432082.1 |
CCDC163P
|
coiled-coil domain containing 163, pseudogene |
| chr20_-_43729750 | 0.01 |
ENST00000537075.1
ENST00000306117.1 |
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| chr2_+_73461364 | 0.01 |
ENST00000540468.1
ENST00000539919.1 ENST00000258091.5 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr3_-_19975665 | 0.01 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr12_+_54718904 | 0.01 |
ENST00000262061.2
ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr1_+_12851545 | 0.01 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr12_-_56727676 | 0.01 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr2_+_73461410 | 0.01 |
ENST00000399032.2
ENST00000398422.2 ENST00000537131.1 ENST00000538797.1 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr7_-_48068643 | 0.00 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr7_+_1727755 | 0.00 |
ENST00000424383.2
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr7_-_135412925 | 0.00 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr17_+_4643300 | 0.00 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr18_+_52258390 | 0.00 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
| chr3_-_16524357 | 0.00 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr2_-_14541060 | 0.00 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr8_+_86999516 | 0.00 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr17_-_12921270 | 0.00 |
ENST00000578071.1
ENST00000426905.3 ENST00000395962.2 ENST00000583371.1 ENST00000338034.4 |
ELAC2
|
elaC ribonuclease Z 2 |
| chrX_-_71792477 | 0.00 |
ENST00000421523.1
ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8
|
histone deacetylase 8 |
| chr3_+_138340049 | 0.00 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of DMBX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) |