Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
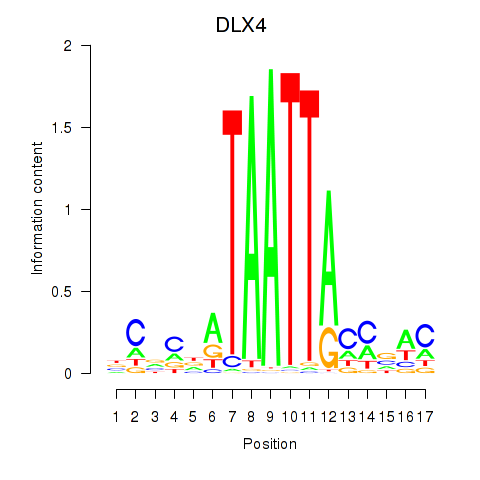
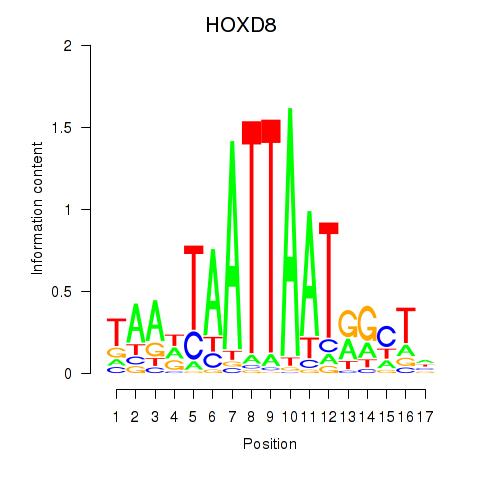
Results for DLX4_HOXD8
Z-value: 0.39
Transcription factors associated with DLX4_HOXD8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX4
|
ENSG00000108813.9 | distal-less homeobox 4 |
|
HOXD8
|
ENSG00000175879.7 | homeobox D8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX4 | hg19_v2_chr17_+_48046538_48046575 | 0.66 | 3.4e-01 | Click! |
| HOXD8 | hg19_v2_chr2_+_176994408_176994641 | 0.59 | 4.1e-01 | Click! |
Activity profile of DLX4_HOXD8 motif
Sorted Z-values of DLX4_HOXD8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_25150373 | 0.18 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr3_-_12587055 | 0.17 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr4_+_9446156 | 0.17 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr1_+_81001398 | 0.16 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr4_-_178169927 | 0.16 |
ENST00000512516.1
|
RP11-487E13.1
|
Uncharacterized protein |
| chr14_-_81425828 | 0.16 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr1_+_81106951 | 0.16 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr21_-_19858196 | 0.15 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr22_+_50981079 | 0.13 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr8_+_82066514 | 0.12 |
ENST00000519412.1
ENST00000521953.1 |
RP11-1149M10.2
|
RP11-1149M10.2 |
| chr17_+_61151306 | 0.12 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr6_-_26027480 | 0.11 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr15_+_75639372 | 0.11 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chrX_-_109590174 | 0.11 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr19_-_58204128 | 0.11 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr13_-_30160925 | 0.10 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr12_-_70093235 | 0.10 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr8_+_101349823 | 0.09 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr1_+_225600404 | 0.09 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr18_+_77905794 | 0.09 |
ENST00000587254.1
ENST00000586421.1 |
AC139100.2
|
Uncharacterized protein |
| chr12_-_118628315 | 0.09 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr16_-_21863541 | 0.09 |
ENST00000543654.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr3_+_186158169 | 0.09 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr12_-_10573149 | 0.08 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr4_-_120243545 | 0.08 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr8_-_66474884 | 0.08 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr6_+_26156551 | 0.08 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr8_+_94752349 | 0.08 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr12_-_86650077 | 0.08 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr11_-_85376121 | 0.07 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr6_-_27841289 | 0.07 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr12_-_24737089 | 0.07 |
ENST00000483544.1
|
LINC00477
|
long intergenic non-protein coding RNA 477 |
| chr6_+_26104104 | 0.07 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chrY_+_15418467 | 0.07 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr6_-_26032288 | 0.07 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr2_-_203735586 | 0.07 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr19_+_41882466 | 0.07 |
ENST00000436170.2
|
TMEM91
|
transmembrane protein 91 |
| chr16_-_25122785 | 0.07 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr7_+_13141010 | 0.07 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr2_+_161993465 | 0.07 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_111649074 | 0.07 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr19_-_58864848 | 0.07 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr7_+_93652116 | 0.07 |
ENST00000415536.1
|
AC003092.1
|
AC003092.1 |
| chr5_+_42756903 | 0.07 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr12_+_78359999 | 0.07 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr5_+_140227357 | 0.07 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr12_+_70574088 | 0.06 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr12_+_14572070 | 0.06 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr4_+_70916119 | 0.06 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr6_+_26045603 | 0.06 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr4_-_48116540 | 0.06 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr1_+_84630574 | 0.06 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_-_85771168 | 0.06 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr4_-_89442940 | 0.06 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr4_+_123653807 | 0.06 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr19_+_42041860 | 0.06 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr6_+_30856507 | 0.06 |
ENST00000513240.1
ENST00000424544.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_+_26204825 | 0.06 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr11_+_5710919 | 0.06 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr15_-_54267147 | 0.05 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr19_-_45457264 | 0.05 |
ENST00000591646.1
|
CTB-129P6.11
|
Uncharacterized protein |
| chr2_+_108994466 | 0.05 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr11_+_3011093 | 0.05 |
ENST00000332881.2
|
AC131971.1
|
HCG1782999; PRO0943; Uncharacterized protein |
| chr1_+_155179012 | 0.05 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr1_-_89591749 | 0.05 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr11_-_102651343 | 0.05 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr19_+_52873166 | 0.05 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr5_+_129083772 | 0.05 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr19_-_16008880 | 0.05 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr4_+_96012585 | 0.05 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr10_-_93669233 | 0.05 |
ENST00000311575.5
|
FGFBP3
|
fibroblast growth factor binding protein 3 |
| chr10_+_57358750 | 0.05 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr19_-_52674896 | 0.05 |
ENST00000322146.8
ENST00000597065.1 |
ZNF836
|
zinc finger protein 836 |
| chr11_+_35222629 | 0.05 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr3_+_25831567 | 0.05 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr12_+_76653682 | 0.05 |
ENST00000553247.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr17_+_67498396 | 0.05 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr15_+_76016293 | 0.05 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr20_+_56964169 | 0.05 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr1_-_116926718 | 0.05 |
ENST00000598661.1
|
AL136376.1
|
Uncharacterized protein |
| chr11_-_72070206 | 0.05 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr19_-_56879416 | 0.05 |
ENST00000591036.1
|
AC006116.21
|
AC006116.21 |
| chr22_-_18923655 | 0.05 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr12_+_133707570 | 0.05 |
ENST00000416488.1
ENST00000540096.2 |
ZNF268
CTD-2140B24.4
|
zinc finger protein 268 Zinc finger protein 268 |
| chr9_-_27005686 | 0.05 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr14_-_21566731 | 0.05 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr16_-_28634874 | 0.05 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr2_+_105050794 | 0.05 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr19_+_1908257 | 0.04 |
ENST00000411971.1
|
SCAMP4
|
secretory carrier membrane protein 4 |
| chr12_-_70093111 | 0.04 |
ENST00000548658.1
ENST00000476098.1 ENST00000331471.4 ENST00000393365.1 |
BEST3
|
bestrophin 3 |
| chr12_-_111142729 | 0.04 |
ENST00000546713.1
|
HVCN1
|
hydrogen voltage-gated channel 1 |
| chr5_-_135290705 | 0.04 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr4_+_75174180 | 0.04 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr5_-_135290651 | 0.04 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr16_+_22517166 | 0.04 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr1_+_149822620 | 0.04 |
ENST00000369159.2
|
HIST2H2AA4
|
histone cluster 2, H2aa4 |
| chr12_-_10282836 | 0.04 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr20_-_32580924 | 0.04 |
ENST00000432859.1
|
RP5-1125A11.1
|
RP5-1125A11.1 |
| chr7_+_80275953 | 0.04 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr6_+_27861190 | 0.04 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr2_+_196440692 | 0.04 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr4_+_40198527 | 0.04 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr13_-_30880979 | 0.04 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr6_-_52668605 | 0.04 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr8_-_25281747 | 0.04 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr3_+_187420101 | 0.04 |
ENST00000449623.1
ENST00000437407.1 |
RP11-211G3.3
|
Uncharacterized protein |
| chr16_-_71323617 | 0.04 |
ENST00000563876.1
|
CMTR2
|
cap methyltransferase 2 |
| chr14_-_91720224 | 0.04 |
ENST00000238699.3
ENST00000531499.2 |
GPR68
|
G protein-coupled receptor 68 |
| chr9_+_131683174 | 0.04 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr4_+_110736659 | 0.04 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr7_-_111032971 | 0.04 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_58176525 | 0.04 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr18_+_3466248 | 0.04 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr8_+_87111059 | 0.04 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr11_+_62496114 | 0.04 |
ENST00000532583.1
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr19_-_48759119 | 0.04 |
ENST00000522889.1
ENST00000520753.1 ENST00000519940.1 ENST00000519332.1 ENST00000521437.1 ENST00000520007.1 ENST00000521613.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr3_+_172034218 | 0.04 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr7_-_105221898 | 0.04 |
ENST00000486180.1
ENST00000485614.1 ENST00000480514.1 |
EFCAB10
|
EF-hand calcium binding domain 10 |
| chr15_+_69857515 | 0.04 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr2_+_182850551 | 0.04 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_-_45140074 | 0.04 |
ENST00000420706.1
ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53
|
transmembrane protein 53 |
| chr6_+_125304502 | 0.04 |
ENST00000519799.1
ENST00000368414.2 ENST00000359704.2 |
RNF217
|
ring finger protein 217 |
| chr11_+_110225855 | 0.04 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr8_-_38008783 | 0.04 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr19_-_47291843 | 0.04 |
ENST00000542575.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr5_-_55412774 | 0.04 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr2_-_134326009 | 0.04 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr3_+_118905564 | 0.04 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr1_+_74701062 | 0.04 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr19_-_52408257 | 0.04 |
ENST00000354957.3
ENST00000600738.1 ENST00000595418.1 ENST00000599530.1 |
ZNF649
|
zinc finger protein 649 |
| chr15_-_78913628 | 0.04 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr14_+_38264418 | 0.04 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr8_+_40018977 | 0.04 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr18_+_52385068 | 0.04 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr6_-_26285737 | 0.04 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr15_+_40697988 | 0.04 |
ENST00000487418.2
ENST00000479013.2 |
IVD
|
isovaleryl-CoA dehydrogenase |
| chr1_-_205091115 | 0.04 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr9_+_22646189 | 0.04 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr11_-_58611957 | 0.04 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr9_-_95055923 | 0.04 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr13_-_30881134 | 0.04 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr19_+_55996316 | 0.04 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr4_-_119759795 | 0.04 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr5_-_119669160 | 0.03 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr12_-_120241187 | 0.03 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr1_-_117021430 | 0.03 |
ENST00000423907.1
ENST00000434879.1 ENST00000443219.1 |
RP4-655J12.4
|
RP4-655J12.4 |
| chr12_+_76653611 | 0.03 |
ENST00000550380.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr1_+_196743912 | 0.03 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr9_-_116837249 | 0.03 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr11_+_92085262 | 0.03 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr11_-_128894053 | 0.03 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr4_+_144312659 | 0.03 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr22_+_20703829 | 0.03 |
ENST00000583722.1
|
FAM230A
|
family with sequence similarity 230, member A |
| chr17_+_43238438 | 0.03 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr8_+_67104323 | 0.03 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr1_-_201140673 | 0.03 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr14_-_107283278 | 0.03 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr6_-_112575758 | 0.03 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr8_-_6735451 | 0.03 |
ENST00000297439.3
|
DEFB1
|
defensin, beta 1 |
| chr10_+_5005445 | 0.03 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr10_-_97200772 | 0.03 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr8_-_124553437 | 0.03 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr14_-_71001708 | 0.03 |
ENST00000256389.3
|
ADAM20
|
ADAM metallopeptidase domain 20 |
| chr1_+_240177627 | 0.03 |
ENST00000447095.1
|
FMN2
|
formin 2 |
| chr12_+_110011571 | 0.03 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr3_+_121289551 | 0.03 |
ENST00000334384.3
|
ARGFX
|
arginine-fifty homeobox |
| chr6_-_26235206 | 0.03 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr11_+_73661364 | 0.03 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr6_-_135271219 | 0.03 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr21_+_43619796 | 0.03 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr4_-_103749205 | 0.03 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_42946947 | 0.03 |
ENST00000304611.8
|
PEX6
|
peroxisomal biogenesis factor 6 |
| chr8_-_37707356 | 0.03 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr7_+_66800928 | 0.03 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr17_+_34171081 | 0.03 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr11_-_47447970 | 0.03 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr3_+_108015382 | 0.03 |
ENST00000463019.3
ENST00000491820.1 ENST00000467562.1 ENST00000482430.1 ENST00000462629.1 |
HHLA2
|
HERV-H LTR-associating 2 |
| chr11_+_2397402 | 0.03 |
ENST00000475945.2
|
CD81
|
CD81 molecule |
| chr20_-_50384864 | 0.03 |
ENST00000311637.5
ENST00000402822.1 |
ATP9A
|
ATPase, class II, type 9A |
| chr7_-_55620433 | 0.03 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr2_-_37501692 | 0.03 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr1_+_197237352 | 0.03 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr7_-_99716914 | 0.03 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_-_73663245 | 0.03 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr14_-_35591156 | 0.03 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr2_-_86333244 | 0.03 |
ENST00000263857.6
ENST00000409681.1 |
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa |
| chr10_-_71169031 | 0.03 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr20_+_3801162 | 0.03 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr10_+_5005598 | 0.03 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr19_+_21203481 | 0.03 |
ENST00000595401.1
|
ZNF430
|
zinc finger protein 430 |
| chr14_-_23426322 | 0.03 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr18_+_68002675 | 0.03 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr14_+_101359265 | 0.03 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr19_+_57901208 | 0.03 |
ENST00000366197.5
ENST00000596282.1 ENST00000597400.1 ENST00000598895.1 ENST00000336128.7 ENST00000596617.1 |
ZNF548
AC003002.6
|
zinc finger protein 548 Uncharacterized protein |
| chr2_+_39117010 | 0.03 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr17_-_41050716 | 0.03 |
ENST00000417193.1
ENST00000301683.3 ENST00000436546.1 ENST00000431109.2 |
LINC00671
|
long intergenic non-protein coding RNA 671 |
| chr21_-_22175341 | 0.03 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr16_+_15489603 | 0.03 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr6_+_116601330 | 0.03 |
ENST00000449314.1
ENST00000453463.1 |
RP1-93H18.1
|
RP1-93H18.1 |
| chr8_+_91803695 | 0.03 |
ENST00000417640.2
|
NECAB1
|
N-terminal EF-hand calcium binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX4_HOXD8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.0 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.0 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.0 | GO:0017143 | insecticide metabolic process(GO:0017143) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.0 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.0 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |