Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
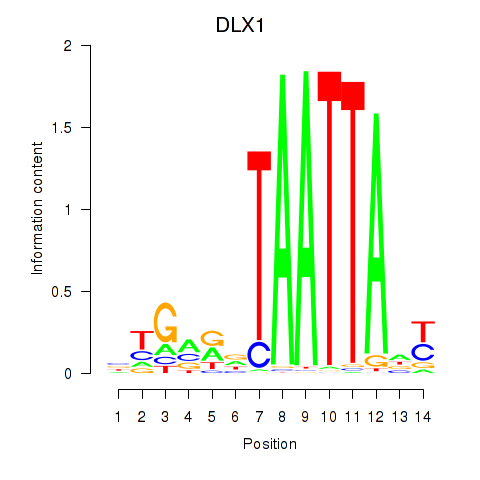
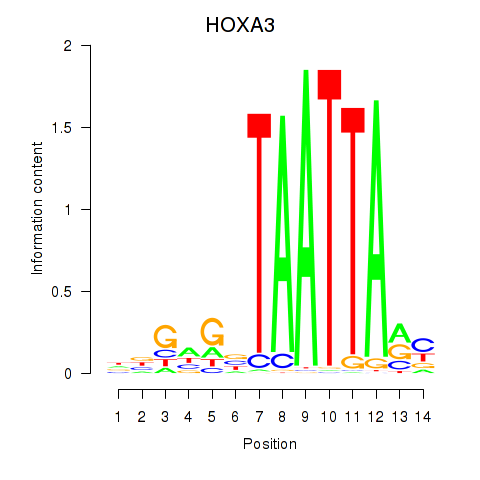
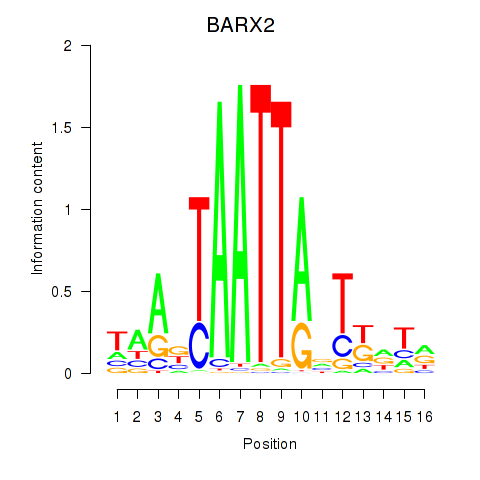
Results for DLX1_HOXA3_BARX2
Z-value: 0.58
Transcription factors associated with DLX1_HOXA3_BARX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX1
|
ENSG00000144355.10 | distal-less homeobox 1 |
|
HOXA3
|
ENSG00000105997.18 | homeobox A3 |
|
BARX2
|
ENSG00000043039.5 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX1 | hg19_v2_chr2_+_172949468_172949522 | 0.69 | 3.1e-01 | Click! |
| HOXA3 | hg19_v2_chr7_-_27179814_27179840 | 0.67 | 3.3e-01 | Click! |
Activity profile of DLX1_HOXA3_BARX2 motif
Sorted Z-values of DLX1_HOXA3_BARX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_115498761 | 0.25 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr17_-_57229155 | 0.23 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr5_+_136070614 | 0.23 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr7_-_99716914 | 0.23 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_+_74846230 | 0.22 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr14_+_56584414 | 0.21 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr11_+_59705928 | 0.19 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr15_+_63188009 | 0.19 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr1_+_54569968 | 0.19 |
ENST00000391366.1
|
AL161915.1
|
Uncharacterized protein |
| chr1_+_212965170 | 0.19 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr3_-_167191814 | 0.19 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr12_-_10978957 | 0.18 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr4_-_185275104 | 0.15 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr4_+_80584903 | 0.15 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr5_-_139726181 | 0.14 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr20_-_29896388 | 0.14 |
ENST00000400549.1
|
DEFB116
|
defensin, beta 116 |
| chr17_-_46690839 | 0.12 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr4_-_130692631 | 0.12 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr9_-_5339873 | 0.12 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr6_+_155537771 | 0.12 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr16_-_25122735 | 0.12 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr10_-_128110441 | 0.12 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr16_+_53412368 | 0.12 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr10_-_28571015 | 0.12 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr2_-_201753980 | 0.12 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr3_-_98235962 | 0.11 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr18_+_59415396 | 0.11 |
ENST00000567801.1
|
RP11-1096D5.1
|
RP11-1096D5.1 |
| chr11_-_13011081 | 0.11 |
ENST00000532541.1
ENST00000526388.1 ENST00000534477.1 ENST00000531402.1 ENST00000527945.1 ENST00000504230.2 |
LINC00958
|
long intergenic non-protein coding RNA 958 |
| chrX_-_11129229 | 0.10 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr4_-_36245561 | 0.10 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr17_-_5321549 | 0.10 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr15_+_92006567 | 0.10 |
ENST00000554333.1
|
RP11-661P17.1
|
RP11-661P17.1 |
| chr14_+_62164340 | 0.10 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr6_-_47445214 | 0.10 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr5_+_142286887 | 0.10 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr12_+_22852791 | 0.10 |
ENST00000413794.2
|
RP11-114G22.1
|
RP11-114G22.1 |
| chr19_+_36239576 | 0.10 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr1_+_160370344 | 0.09 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr7_+_116595028 | 0.09 |
ENST00000397751.1
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr4_-_74486109 | 0.09 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_+_30589829 | 0.09 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr8_+_107738343 | 0.09 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr13_+_78315295 | 0.09 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chrX_+_43515467 | 0.09 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr2_+_228736335 | 0.09 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr16_+_15489629 | 0.09 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr6_+_160693591 | 0.09 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr3_-_123680246 | 0.09 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr4_+_95128748 | 0.09 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_66536248 | 0.09 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr12_+_52695617 | 0.09 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr9_+_130911723 | 0.08 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr1_-_207226313 | 0.08 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr7_-_64023441 | 0.08 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr1_-_22215192 | 0.08 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr14_+_67831576 | 0.08 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr4_-_69536346 | 0.08 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr5_+_53686658 | 0.08 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr15_+_64680003 | 0.08 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr6_-_26235206 | 0.08 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr14_+_85994943 | 0.08 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr19_-_36606181 | 0.08 |
ENST00000221859.4
|
POLR2I
|
polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa |
| chr4_-_112993808 | 0.08 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr10_-_115904361 | 0.08 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chrM_+_8366 | 0.07 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr12_-_8088773 | 0.07 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr7_-_150020814 | 0.07 |
ENST00000477871.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr11_+_94706973 | 0.07 |
ENST00000536741.1
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chrX_+_108779004 | 0.07 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr7_-_124569991 | 0.07 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr8_-_41166953 | 0.07 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr7_-_24957699 | 0.07 |
ENST00000441059.1
ENST00000415162.1 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr17_-_47786375 | 0.07 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr2_+_234826016 | 0.07 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr10_+_5238793 | 0.07 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr1_-_242612726 | 0.07 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr7_-_44580861 | 0.06 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chrX_-_16887963 | 0.06 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr12_-_76461249 | 0.06 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr11_+_24518723 | 0.06 |
ENST00000336930.6
ENST00000529015.1 ENST00000533227.1 |
LUZP2
|
leucine zipper protein 2 |
| chrX_-_1331527 | 0.06 |
ENST00000381567.3
ENST00000381566.1 ENST00000400841.2 |
CRLF2
|
cytokine receptor-like factor 2 |
| chr6_-_138866823 | 0.06 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr8_-_30670384 | 0.06 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr6_-_32908765 | 0.06 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr18_-_33709268 | 0.06 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr12_-_12837423 | 0.06 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr7_-_14029283 | 0.06 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr2_+_191792376 | 0.06 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr12_-_25150409 | 0.06 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr1_+_109265033 | 0.06 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr14_+_32798547 | 0.06 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr2_-_201753859 | 0.06 |
ENST00000409361.1
ENST00000392283.4 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr1_-_242612779 | 0.06 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr13_+_78315348 | 0.06 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr10_-_135379132 | 0.06 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr15_+_35270552 | 0.06 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr2_-_203735484 | 0.06 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr1_+_43613566 | 0.06 |
ENST00000409396.1
|
FAM183A
|
family with sequence similarity 183, member A |
| chr15_-_99057551 | 0.06 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr1_+_68150744 | 0.06 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr9_+_131062367 | 0.06 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr3_-_180397256 | 0.06 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr2_-_151395525 | 0.06 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr2_+_26624775 | 0.06 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr15_-_54267147 | 0.05 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr5_+_174151536 | 0.05 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr9_+_125133315 | 0.05 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr14_+_57671888 | 0.05 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr14_-_36988882 | 0.05 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr2_-_74618907 | 0.05 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr6_+_13272904 | 0.05 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr7_+_116660246 | 0.05 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr11_+_29181503 | 0.05 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr3_+_152552685 | 0.05 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr3_-_101039402 | 0.05 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr10_+_695888 | 0.05 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr19_-_51920952 | 0.05 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr19_+_21688366 | 0.05 |
ENST00000358491.4
ENST00000597078.1 |
ZNF429
|
zinc finger protein 429 |
| chr12_-_96336369 | 0.05 |
ENST00000546947.1
ENST00000546386.1 |
CCDC38
|
coiled-coil domain containing 38 |
| chr2_+_87808725 | 0.05 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr6_+_153552455 | 0.05 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr14_-_54425475 | 0.05 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr11_+_46193466 | 0.05 |
ENST00000533793.1
|
RP11-702F3.3
|
RP11-702F3.3 |
| chr10_+_35484793 | 0.05 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr12_-_10282742 | 0.05 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr3_-_20053741 | 0.05 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr4_-_109541539 | 0.05 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chrX_-_23926004 | 0.05 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr2_-_169887827 | 0.05 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr14_-_94789663 | 0.05 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr16_+_31885079 | 0.05 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr5_+_66300464 | 0.05 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_212965104 | 0.05 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr11_-_117748138 | 0.05 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr7_-_150020750 | 0.05 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr3_-_48659193 | 0.05 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr12_+_20963632 | 0.05 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr17_-_57158523 | 0.05 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr16_+_15489603 | 0.05 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr19_+_21579908 | 0.05 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr12_-_96794330 | 0.05 |
ENST00000261211.3
|
CDK17
|
cyclin-dependent kinase 17 |
| chr15_+_84115868 | 0.05 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr11_-_327537 | 0.05 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chrM_+_8527 | 0.05 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr3_+_46618727 | 0.05 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr15_-_101835110 | 0.05 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr2_-_201753717 | 0.05 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr7_-_92777606 | 0.05 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr2_+_219125714 | 0.05 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr1_-_109618566 | 0.04 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr9_+_130911770 | 0.04 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr8_-_91618285 | 0.04 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr6_-_31689456 | 0.04 |
ENST00000495859.1
ENST00000375819.2 |
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C |
| chr14_-_23426231 | 0.04 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr12_-_48164812 | 0.04 |
ENST00000549151.1
ENST00000548919.1 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr19_-_58662139 | 0.04 |
ENST00000598312.1
|
ZNF329
|
zinc finger protein 329 |
| chr11_+_60552797 | 0.04 |
ENST00000308287.1
|
MS4A10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chr15_+_67390920 | 0.04 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr12_+_20963647 | 0.04 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr17_-_8263538 | 0.04 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr17_-_73901494 | 0.04 |
ENST00000309352.3
|
MRPL38
|
mitochondrial ribosomal protein L38 |
| chr6_-_138833630 | 0.04 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr1_-_222763101 | 0.04 |
ENST00000391883.2
ENST00000366890.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr18_-_46895066 | 0.04 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr7_-_14028488 | 0.04 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr11_-_117747607 | 0.04 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr2_+_201450591 | 0.04 |
ENST00000374700.2
|
AOX1
|
aldehyde oxidase 1 |
| chr12_+_51818749 | 0.04 |
ENST00000514353.3
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr10_+_90660832 | 0.04 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr12_+_109490370 | 0.04 |
ENST00000257548.5
ENST00000536723.1 ENST00000536393.1 |
USP30
|
ubiquitin specific peptidase 30 |
| chr12_-_123450986 | 0.04 |
ENST00000344275.7
ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr1_+_207277590 | 0.04 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr17_+_48823896 | 0.04 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr14_-_36645674 | 0.04 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr6_-_170151603 | 0.04 |
ENST00000366774.3
|
TCTE3
|
t-complex-associated-testis-expressed 3 |
| chr12_-_112123524 | 0.04 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr2_+_201754050 | 0.04 |
ENST00000426253.1
ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr19_+_21324827 | 0.04 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr22_-_32766972 | 0.04 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr3_-_108248169 | 0.04 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr14_+_96000930 | 0.04 |
ENST00000331334.4
|
GLRX5
|
glutaredoxin 5 |
| chr22_-_22863466 | 0.04 |
ENST00000406426.1
ENST00000360412.2 |
ZNF280B
|
zinc finger protein 280B |
| chrX_-_130423200 | 0.04 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr10_-_135379074 | 0.04 |
ENST00000303903.6
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr17_-_48785216 | 0.04 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr15_-_54051831 | 0.04 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr3_+_98482175 | 0.04 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr12_-_50616382 | 0.04 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr9_+_135937365 | 0.04 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr17_-_39623681 | 0.04 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr16_+_28889801 | 0.04 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr17_+_59489112 | 0.04 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr17_-_64225508 | 0.04 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr19_+_45417921 | 0.04 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr19_-_35264089 | 0.04 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chr2_-_201729284 | 0.04 |
ENST00000434813.2
|
CLK1
|
CDC-like kinase 1 |
| chr14_-_45252031 | 0.03 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr20_+_18488137 | 0.03 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr5_-_96478466 | 0.03 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
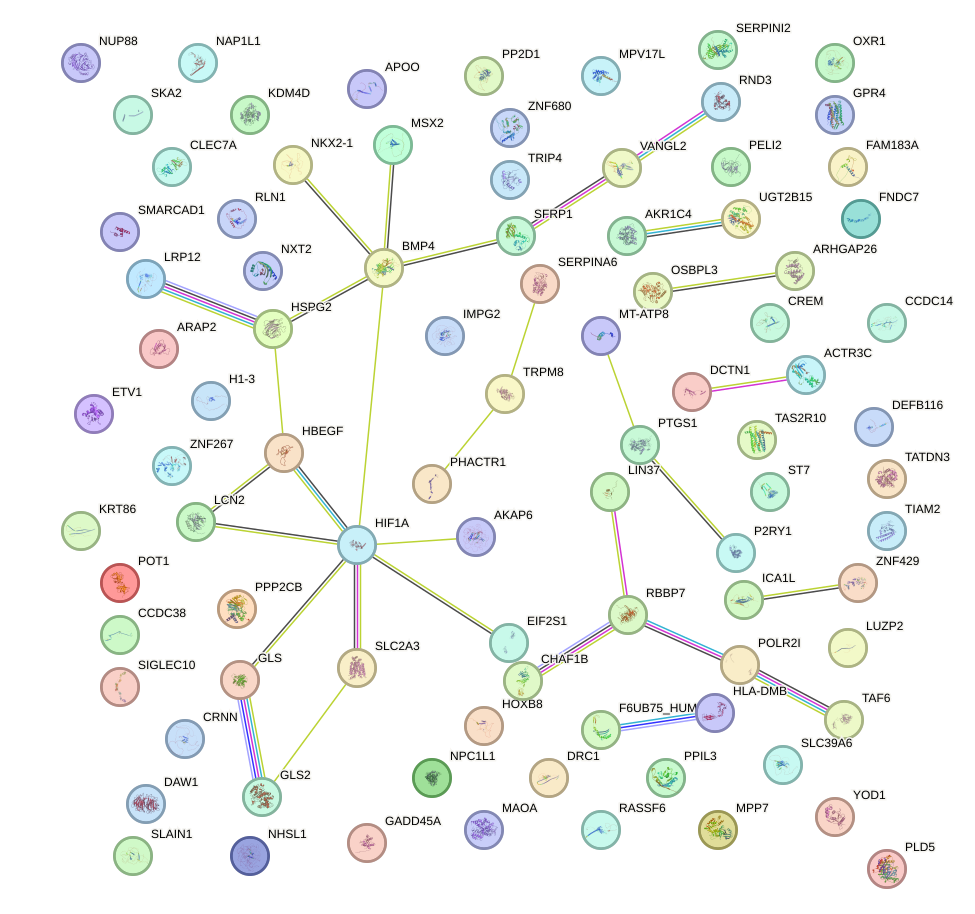
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX1_HOXA3_BARX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0090244 | negative regulation of B cell differentiation(GO:0045578) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0060405 | regulation of penile erection(GO:0060405) |
| 0.0 | 0.1 | GO:1905072 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.0 | GO:2001190 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.0 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.0 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.0 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.0 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |