Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
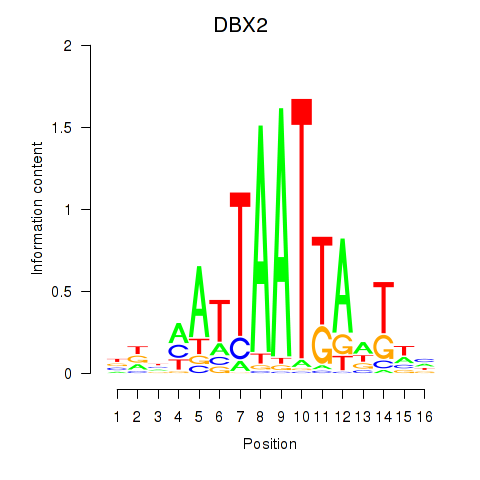
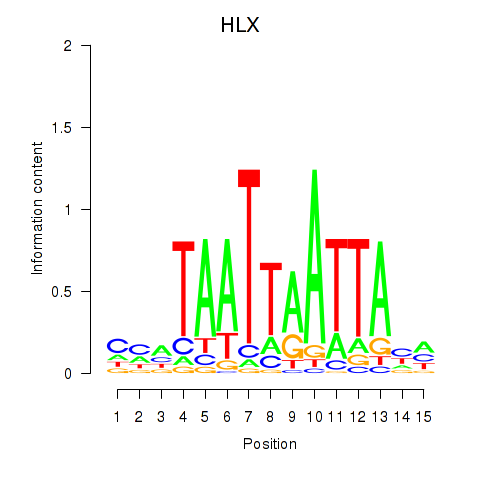
Results for DBX2_HLX
Z-value: 0.57
Transcription factors associated with DBX2_HLX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBX2
|
ENSG00000185610.6 | developing brain homeobox 2 |
|
HLX
|
ENSG00000136630.11 | H2.0 like homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLX | hg19_v2_chr1_+_221051699_221051699 | 0.88 | 1.2e-01 | Click! |
Activity profile of DBX2_HLX motif
Sorted Z-values of DBX2_HLX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_10978957 | 0.44 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr21_+_33671264 | 0.37 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr11_+_5710919 | 0.36 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr16_+_12059091 | 0.35 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr8_-_145754428 | 0.32 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr9_-_21482312 | 0.26 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr1_+_117963209 | 0.25 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr7_+_138915102 | 0.25 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr17_+_61151306 | 0.24 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr19_-_3557570 | 0.23 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr4_-_89442940 | 0.19 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr22_-_39268308 | 0.15 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr12_-_85430024 | 0.14 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr19_+_50016610 | 0.14 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_-_327537 | 0.14 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr2_+_135596180 | 0.14 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr19_+_50016411 | 0.14 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr16_-_66583994 | 0.13 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr1_-_183538319 | 0.13 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr3_-_114790179 | 0.12 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr13_-_45048386 | 0.12 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr1_+_76251912 | 0.12 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr19_-_22018966 | 0.12 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr5_+_175288631 | 0.12 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr6_+_30029008 | 0.12 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr22_+_30821732 | 0.12 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr6_-_25874440 | 0.12 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr5_+_102200948 | 0.12 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_+_138340049 | 0.11 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr7_+_13141010 | 0.11 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chrM_-_14670 | 0.11 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr4_+_71019903 | 0.11 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr4_+_147096837 | 0.11 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr16_-_66584059 | 0.11 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr16_-_28937027 | 0.11 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr2_+_135596106 | 0.11 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr7_-_44122063 | 0.10 |
ENST00000335195.6
ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM
|
polymerase (DNA directed), mu |
| chr10_-_22292675 | 0.10 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr11_+_327171 | 0.10 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chrM_+_8366 | 0.10 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr19_-_58864848 | 0.10 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chrX_-_77225135 | 0.10 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr17_+_17685422 | 0.09 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr2_-_211342292 | 0.09 |
ENST00000448951.1
|
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr6_-_47445214 | 0.09 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr9_-_3469181 | 0.09 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr4_+_87857538 | 0.09 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr10_+_91152303 | 0.09 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr1_-_89591749 | 0.09 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr9_+_67977438 | 0.09 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr2_+_161993412 | 0.09 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr11_+_45825616 | 0.09 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr1_+_221051699 | 0.09 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr1_-_108743471 | 0.08 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr11_-_47521309 | 0.08 |
ENST00000535982.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr12_+_14572070 | 0.08 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr11_-_559377 | 0.08 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr2_-_203735484 | 0.08 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr14_-_54425475 | 0.08 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr6_+_114178512 | 0.08 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr2_+_208414985 | 0.08 |
ENST00000414681.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr18_+_61445205 | 0.08 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr12_+_21207503 | 0.07 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr9_+_131549610 | 0.07 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr8_+_120885949 | 0.07 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr1_+_196788887 | 0.07 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr21_-_16125773 | 0.07 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr2_-_183106641 | 0.07 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr16_-_28634874 | 0.07 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr9_-_95055923 | 0.07 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr8_-_124741451 | 0.07 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr8_-_112039643 | 0.07 |
ENST00000524283.1
|
RP11-946L20.2
|
RP11-946L20.2 |
| chr1_-_204183071 | 0.07 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr15_+_67418047 | 0.07 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr1_-_168464875 | 0.07 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr5_+_157158205 | 0.06 |
ENST00000231198.7
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr11_+_5711010 | 0.06 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr14_+_52164675 | 0.06 |
ENST00000555936.1
|
FRMD6
|
FERM domain containing 6 |
| chr16_-_29934558 | 0.06 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr17_+_74463650 | 0.06 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr6_+_26087509 | 0.06 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr22_+_23487513 | 0.06 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr20_-_33735070 | 0.06 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr17_-_7760457 | 0.06 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr5_-_175388327 | 0.06 |
ENST00000432305.2
ENST00000505969.1 |
THOC3
|
THO complex 3 |
| chr13_-_110438914 | 0.06 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr11_+_44117219 | 0.06 |
ENST00000532479.1
ENST00000527014.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr1_+_160370344 | 0.06 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr12_+_57623477 | 0.06 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_+_151253991 | 0.06 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr2_-_37068530 | 0.06 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr10_-_63995871 | 0.06 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr11_+_100784231 | 0.06 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr7_-_92777606 | 0.06 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr10_-_53459319 | 0.06 |
ENST00000331173.4
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
| chr22_+_46476192 | 0.06 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chrX_+_150565038 | 0.06 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr1_-_110950564 | 0.06 |
ENST00000256644.4
|
LAMTOR5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr11_-_111649015 | 0.06 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chrX_+_55744228 | 0.06 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr16_-_21875424 | 0.05 |
ENST00000541674.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr14_-_67826538 | 0.05 |
ENST00000553687.1
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr4_-_40477766 | 0.05 |
ENST00000507180.1
|
RBM47
|
RNA binding motif protein 47 |
| chr8_-_133637624 | 0.05 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr2_-_87248975 | 0.05 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chrM_+_10464 | 0.05 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr1_+_149239529 | 0.05 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr4_+_186990298 | 0.05 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chrX_+_9502971 | 0.05 |
ENST00000452824.1
|
TBL1X
|
transducin (beta)-like 1X-linked |
| chr2_+_201987200 | 0.05 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr17_+_67759813 | 0.05 |
ENST00000587241.1
|
AC003051.1
|
AC003051.1 |
| chr8_-_42396185 | 0.05 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr1_-_89357179 | 0.05 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chrM_+_10053 | 0.05 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr3_-_150421752 | 0.05 |
ENST00000498386.1
|
FAM194A
|
family with sequence similarity 194, member A |
| chr14_+_35591509 | 0.05 |
ENST00000604073.1
|
KIAA0391
|
KIAA0391 |
| chr9_+_131549483 | 0.05 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr5_+_43602750 | 0.05 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr22_-_50970919 | 0.05 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr4_+_26324474 | 0.05 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_-_177116772 | 0.05 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr1_+_225600404 | 0.05 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr8_-_116504448 | 0.05 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr7_-_22862406 | 0.05 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr3_-_112564797 | 0.05 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr5_-_33297946 | 0.05 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
| chr11_-_64684672 | 0.05 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr8_+_109455830 | 0.05 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr2_+_120687335 | 0.05 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr2_+_234580499 | 0.05 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr1_-_160549235 | 0.05 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr6_+_34204642 | 0.05 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr12_+_9066472 | 0.05 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr3_-_148939598 | 0.05 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr15_-_99789736 | 0.05 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr5_-_146781153 | 0.05 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr17_+_29664830 | 0.05 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr17_+_8191815 | 0.04 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr2_+_74685413 | 0.04 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr2_-_70520539 | 0.04 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr11_+_110300607 | 0.04 |
ENST00000260270.2
|
FDX1
|
ferredoxin 1 |
| chr16_-_66583701 | 0.04 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr10_+_696000 | 0.04 |
ENST00000381489.5
|
PRR26
|
proline rich 26 |
| chr11_-_104827425 | 0.04 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_+_87135076 | 0.04 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr9_-_5833027 | 0.04 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr18_+_61616510 | 0.04 |
ENST00000408945.3
|
HMSD
|
histocompatibility (minor) serpin domain containing |
| chr14_-_23292596 | 0.04 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr10_+_32873190 | 0.04 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr2_-_228582709 | 0.04 |
ENST00000541617.1
ENST00000409456.2 ENST00000409287.1 ENST00000258403.3 |
SLC19A3
|
solute carrier family 19 (thiamine transporter), member 3 |
| chr4_-_186570679 | 0.04 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_-_154689596 | 0.04 |
ENST00000369444.2
|
H2AFB3
|
H2A histone family, member B3 |
| chr15_+_22368478 | 0.04 |
ENST00000332663.2
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2 |
| chr19_-_36304201 | 0.04 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr7_+_13141097 | 0.04 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr15_-_64385981 | 0.04 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr3_-_141747950 | 0.04 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_-_26593649 | 0.04 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr5_-_149829294 | 0.04 |
ENST00000401695.3
|
RPS14
|
ribosomal protein S14 |
| chr1_+_196743943 | 0.04 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr1_+_81771806 | 0.04 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr14_-_101295407 | 0.04 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr7_+_141811539 | 0.04 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr4_+_89513574 | 0.04 |
ENST00000402738.1
ENST00000431413.1 ENST00000422770.1 ENST00000407637.1 |
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr2_+_11696464 | 0.04 |
ENST00000234142.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr7_+_23719749 | 0.04 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr4_+_159131630 | 0.04 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr13_+_111855399 | 0.04 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr7_-_16844611 | 0.04 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr1_-_54879140 | 0.04 |
ENST00000525990.1
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr17_-_6524159 | 0.04 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr7_-_107883678 | 0.04 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr17_-_72772462 | 0.04 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr1_-_113160826 | 0.04 |
ENST00000538187.1
ENST00000369664.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr5_+_140762268 | 0.04 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr1_-_211307404 | 0.04 |
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr6_+_26087646 | 0.04 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr5_+_140254884 | 0.04 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr13_-_46742630 | 0.04 |
ENST00000416500.1
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr21_+_33671160 | 0.04 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr2_+_234580525 | 0.04 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr4_+_155484155 | 0.04 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr8_-_143859197 | 0.03 |
ENST00000395192.2
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr4_-_159080806 | 0.03 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr12_+_14369524 | 0.03 |
ENST00000538329.1
|
RP11-134N1.2
|
RP11-134N1.2 |
| chr3_-_145940214 | 0.03 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr2_+_170440844 | 0.03 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr1_+_76251879 | 0.03 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr4_+_119809984 | 0.03 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr12_+_26348429 | 0.03 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr7_-_55620433 | 0.03 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr4_-_164534657 | 0.03 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr10_-_38146482 | 0.03 |
ENST00000374648.3
|
ZNF248
|
zinc finger protein 248 |
| chr4_-_74486109 | 0.03 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr12_-_11287243 | 0.03 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr2_+_105050794 | 0.03 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr4_+_113568207 | 0.03 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr2_+_88047606 | 0.03 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr6_+_34725263 | 0.03 |
ENST00000374018.1
ENST00000374017.3 |
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr6_+_26402517 | 0.03 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DBX2_HLX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.3 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0061151 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.0 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.0 | GO:0000806 | Y chromosome(GO:0000806) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031780 | melanocortin receptor binding(GO:0031779) corticotropin hormone receptor binding(GO:0031780) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.3 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |