Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
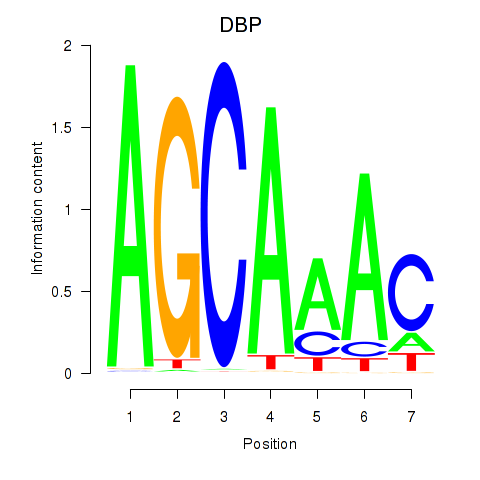
Results for DBP
Z-value: 0.89
Transcription factors associated with DBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBP
|
ENSG00000105516.6 | D-box binding PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DBP | hg19_v2_chr19_-_49140692_49140709 | 0.83 | 1.7e-01 | Click! |
Activity profile of DBP motif
Sorted Z-values of DBP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_37051798 | 0.81 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr16_+_30675654 | 0.60 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr7_+_13141010 | 0.58 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr5_-_27038683 | 0.53 |
ENST00000511822.1
ENST00000231021.4 |
CDH9
|
cadherin 9, type 2 (T1-cadherin) |
| chr11_+_394145 | 0.49 |
ENST00000528036.1
|
PKP3
|
plakophilin 3 |
| chr19_-_56047661 | 0.45 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr16_+_2059872 | 0.43 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr6_-_33168391 | 0.41 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr19_-_41220957 | 0.39 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr6_+_26045603 | 0.39 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr10_+_104178946 | 0.38 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr6_+_26251835 | 0.37 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr2_-_175202151 | 0.37 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr11_-_47206965 | 0.34 |
ENST00000525725.1
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr17_-_56492989 | 0.33 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr21_+_17791648 | 0.33 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_-_118943930 | 0.33 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr17_+_67498396 | 0.31 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_+_149877440 | 0.31 |
ENST00000518299.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr3_-_47023455 | 0.29 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr17_+_72426891 | 0.29 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chrX_+_10031499 | 0.29 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr14_-_69261310 | 0.27 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr6_+_31514622 | 0.27 |
ENST00000376146.4
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr16_+_86612112 | 0.27 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr6_+_100054606 | 0.26 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr2_-_190044480 | 0.26 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr22_+_42196666 | 0.26 |
ENST00000402061.3
ENST00000255784.5 |
CCDC134
|
coiled-coil domain containing 134 |
| chr13_+_44596471 | 0.25 |
ENST00000592085.1
ENST00000591799.1 |
LINC00284
|
long intergenic non-protein coding RNA 284 |
| chr6_-_117747015 | 0.25 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr6_+_21593972 | 0.24 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr6_+_33168637 | 0.24 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr7_+_111846741 | 0.24 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr16_+_85204882 | 0.24 |
ENST00000574293.1
|
CTC-786C10.1
|
CTC-786C10.1 |
| chr6_+_33168597 | 0.23 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr7_-_150946015 | 0.23 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr7_-_65113280 | 0.23 |
ENST00000593865.1
|
AC104057.1
|
Protein LOC100996407 |
| chr1_+_155051379 | 0.23 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr2_+_95963052 | 0.22 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chr11_+_34999328 | 0.22 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr8_+_99076750 | 0.21 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr19_+_36208877 | 0.21 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr9_+_139886846 | 0.21 |
ENST00000371620.3
|
C9orf142
|
chromosome 9 open reading frame 142 |
| chr14_-_21562648 | 0.21 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr9_+_118916082 | 0.21 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr17_-_46667628 | 0.21 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr9_+_35829208 | 0.20 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr17_-_7232585 | 0.20 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr3_-_47950745 | 0.20 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr1_+_61547894 | 0.19 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr11_-_62414070 | 0.19 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr12_-_57030096 | 0.18 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr11_+_394196 | 0.18 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr22_+_31742875 | 0.18 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr7_+_148892557 | 0.18 |
ENST00000262085.3
|
ZNF282
|
zinc finger protein 282 |
| chr16_+_30960375 | 0.17 |
ENST00000318663.4
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr4_-_40477766 | 0.17 |
ENST00000507180.1
|
RBM47
|
RNA binding motif protein 47 |
| chr7_-_135612198 | 0.17 |
ENST00000589735.1
|
LUZP6
|
leucine zipper protein 6 |
| chr2_+_105050794 | 0.17 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr4_-_176733897 | 0.17 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr17_-_46667594 | 0.17 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr17_+_26369865 | 0.17 |
ENST00000582037.1
|
NLK
|
nemo-like kinase |
| chr7_+_142374104 | 0.17 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2-like 6 |
| chr5_+_129083772 | 0.17 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr8_+_107738343 | 0.17 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr16_-_75282088 | 0.16 |
ENST00000542031.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr19_+_13135386 | 0.16 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr14_-_21562671 | 0.16 |
ENST00000554923.1
|
ZNF219
|
zinc finger protein 219 |
| chr17_+_67498295 | 0.15 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr15_+_67418047 | 0.15 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr2_-_208031943 | 0.15 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr15_+_90544532 | 0.14 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr7_+_13141097 | 0.14 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr1_+_155051305 | 0.14 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr14_+_51026844 | 0.14 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr9_-_139372141 | 0.14 |
ENST00000313050.7
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr1_-_151148442 | 0.14 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr1_-_108231101 | 0.13 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr8_-_623547 | 0.13 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr12_+_56511943 | 0.13 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr17_+_2240775 | 0.13 |
ENST00000268989.3
ENST00000426855.2 |
SGSM2
|
small G protein signaling modulator 2 |
| chr16_-_2059748 | 0.13 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr1_+_154229547 | 0.13 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr4_+_113970772 | 0.13 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chrX_-_54070388 | 0.12 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr6_+_88054530 | 0.12 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr5_-_158526756 | 0.12 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr19_+_13134772 | 0.12 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr19_-_48894104 | 0.12 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr16_-_2059797 | 0.12 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr8_+_42195972 | 0.12 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr14_+_24590560 | 0.11 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr19_+_11455900 | 0.11 |
ENST00000588790.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr7_+_114055052 | 0.11 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr17_+_79953310 | 0.11 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr17_-_34308524 | 0.11 |
ENST00000293275.3
|
CCL16
|
chemokine (C-C motif) ligand 16 |
| chr6_-_31514516 | 0.11 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr14_+_51026926 | 0.11 |
ENST00000557735.1
|
ATL1
|
atlastin GTPase 1 |
| chr18_-_2982869 | 0.11 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr10_-_69991865 | 0.11 |
ENST00000373673.3
|
ATOH7
|
atonal homolog 7 (Drosophila) |
| chr19_+_6361440 | 0.11 |
ENST00000245816.4
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr6_+_30687978 | 0.11 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr5_+_140710061 | 0.11 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr17_+_7487146 | 0.11 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr5_+_137673945 | 0.11 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr6_-_18122843 | 0.10 |
ENST00000340650.3
|
NHLRC1
|
NHL repeat containing E3 ubiquitin protein ligase 1 |
| chr17_+_29664830 | 0.10 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr4_-_105416039 | 0.10 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr7_-_104909435 | 0.10 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr2_+_168725458 | 0.10 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr6_+_90272488 | 0.10 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr3_-_48471454 | 0.10 |
ENST00000296440.6
ENST00000448774.2 |
PLXNB1
|
plexin B1 |
| chr1_+_12538594 | 0.09 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_+_167298281 | 0.09 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr5_-_179233314 | 0.09 |
ENST00000523108.1
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chrX_-_124097620 | 0.09 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr17_+_48624450 | 0.09 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr11_+_35684288 | 0.09 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr16_+_67022633 | 0.09 |
ENST00000398354.1
ENST00000326686.5 |
CES4A
|
carboxylesterase 4A |
| chr9_+_2017063 | 0.09 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_-_6420759 | 0.09 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr5_+_179233376 | 0.09 |
ENST00000376929.3
ENST00000514093.1 |
SQSTM1
|
sequestosome 1 |
| chr19_-_42806919 | 0.08 |
ENST00000595530.1
ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr14_+_76452090 | 0.08 |
ENST00000314067.6
ENST00000238628.6 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas) |
| chr11_+_64009072 | 0.08 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr22_+_45714301 | 0.08 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr20_-_22559211 | 0.08 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr8_+_107738240 | 0.08 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr19_+_39881951 | 0.08 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr5_+_32710736 | 0.08 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr9_+_116263778 | 0.08 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr19_+_41856816 | 0.08 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr4_-_23891658 | 0.08 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr9_+_2158443 | 0.08 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_129592471 | 0.08 |
ENST00000473814.2
ENST00000490974.1 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr16_+_67022582 | 0.08 |
ENST00000541479.1
ENST00000338718.4 |
CES4A
|
carboxylesterase 4A |
| chr3_-_171489085 | 0.08 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chrX_+_142967173 | 0.07 |
ENST00000370494.1
|
UBE2NL
|
ubiquitin-conjugating enzyme E2N-like |
| chr1_+_206623784 | 0.07 |
ENST00000426388.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr1_-_85870177 | 0.07 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr19_-_57988871 | 0.07 |
ENST00000596831.1
ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9
ZNF772
|
Uncharacterized protein zinc finger protein 772 |
| chr17_+_42429493 | 0.07 |
ENST00000586242.1
|
GRN
|
granulin |
| chr12_-_92536433 | 0.07 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr8_-_22550815 | 0.07 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr4_-_40516560 | 0.07 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr8_+_85095769 | 0.07 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chrX_+_153769409 | 0.07 |
ENST00000440286.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr8_-_74495065 | 0.07 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr16_+_67022476 | 0.07 |
ENST00000540947.2
|
CES4A
|
carboxylesterase 4A |
| chr11_+_111411384 | 0.07 |
ENST00000375615.3
ENST00000525126.1 ENST00000436913.2 ENST00000533265.1 |
LAYN
|
layilin |
| chr1_+_61548225 | 0.07 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr18_-_56985776 | 0.07 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr1_-_23886285 | 0.07 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr10_-_79397479 | 0.07 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chrX_+_153769446 | 0.07 |
ENST00000422680.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr3_+_141105705 | 0.07 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr10_+_24755416 | 0.07 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr2_-_111291587 | 0.07 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr17_+_48823896 | 0.06 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr17_-_48546232 | 0.06 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chr3_-_112564797 | 0.06 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr5_-_41510656 | 0.06 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr16_+_29789561 | 0.06 |
ENST00000400752.4
|
ZG16
|
zymogen granule protein 16 |
| chr2_-_216257849 | 0.06 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr22_-_31742218 | 0.06 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr14_+_23654525 | 0.06 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr3_-_196910477 | 0.06 |
ENST00000447466.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chrX_+_149531524 | 0.06 |
ENST00000370401.2
|
MAMLD1
|
mastermind-like domain containing 1 |
| chr12_+_56137064 | 0.06 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr20_+_9146969 | 0.05 |
ENST00000416836.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr1_-_173176452 | 0.05 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr6_+_44194762 | 0.05 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr17_+_61271355 | 0.05 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr10_-_79397202 | 0.05 |
ENST00000372437.1
ENST00000372408.2 ENST00000372403.4 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr18_+_18943554 | 0.05 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr5_+_140588269 | 0.05 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr20_+_36405665 | 0.05 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chrX_+_49832231 | 0.05 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr8_+_99076509 | 0.05 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr17_+_1959369 | 0.05 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
hypermethylated in cancer 1 |
| chr8_+_85095553 | 0.05 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr10_-_79397316 | 0.05 |
ENST00000372421.5
ENST00000457953.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_-_60780607 | 0.05 |
ENST00000537768.1
ENST00000335712.6 ENST00000356842.4 |
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr4_-_186732048 | 0.05 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_+_67226019 | 0.05 |
ENST00000379378.3
|
E2F4
|
E2F transcription factor 4, p107/p130-binding |
| chr13_+_93879085 | 0.05 |
ENST00000377047.4
|
GPC6
|
glypican 6 |
| chr18_-_51750948 | 0.05 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr17_-_34195889 | 0.05 |
ENST00000311880.2
|
C17orf66
|
chromosome 17 open reading frame 66 |
| chr15_+_58702742 | 0.05 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr17_-_34195862 | 0.05 |
ENST00000592980.1
ENST00000587626.1 |
C17orf66
|
chromosome 17 open reading frame 66 |
| chr9_+_79634571 | 0.05 |
ENST00000376708.1
|
FOXB2
|
forkhead box B2 |
| chr9_-_13165457 | 0.05 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr20_+_34020827 | 0.04 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr9_-_139371533 | 0.04 |
ENST00000290037.6
ENST00000431893.2 ENST00000371706.3 |
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr13_-_67802549 | 0.04 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr7_+_133615169 | 0.04 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr17_+_2240916 | 0.04 |
ENST00000574563.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr19_+_19496624 | 0.04 |
ENST00000494516.2
ENST00000360315.3 ENST00000252577.5 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr8_-_29120580 | 0.04 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr17_+_38498594 | 0.04 |
ENST00000394081.3
|
RARA
|
retinoic acid receptor, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of DBP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.7 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.2 | GO:0035910 | noradrenergic neuron differentiation(GO:0003357) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.3 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.4 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:2000525 | response to nematode(GO:0009624) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.0 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.2 | GO:1902083 | cellular response to hydroperoxide(GO:0071447) negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |