Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
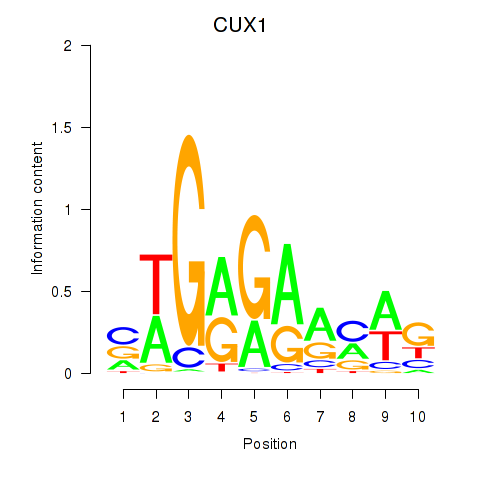
Results for CUX1
Z-value: 0.32
Transcription factors associated with CUX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX1
|
ENSG00000257923.5 | cut like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX1 | hg19_v2_chr7_+_101917407_101917445 | 0.79 | 2.1e-01 | Click! |
Activity profile of CUX1 motif
Sorted Z-values of CUX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_51152702 | 0.18 |
ENST00000425202.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr11_+_35222629 | 0.15 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_-_9482010 | 0.14 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr1_-_153513170 | 0.12 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr14_-_24732738 | 0.10 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr19_-_41220957 | 0.10 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr7_+_141478242 | 0.09 |
ENST00000247881.2
|
TAS2R4
|
taste receptor, type 2, member 4 |
| chr3_-_134093738 | 0.09 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr16_-_776431 | 0.09 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr7_-_99716914 | 0.09 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr8_-_123706338 | 0.08 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr12_+_7072354 | 0.08 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr2_-_130956006 | 0.08 |
ENST00000312988.7
|
TUBA3E
|
tubulin, alpha 3e |
| chr11_+_63742050 | 0.08 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr14_+_21359558 | 0.08 |
ENST00000304639.3
|
RNASE3
|
ribonuclease, RNase A family, 3 |
| chr9_-_107754034 | 0.08 |
ENST00000457720.1
|
RP11-217B7.3
|
RP11-217B7.3 |
| chr16_+_30935896 | 0.07 |
ENST00000562319.1
ENST00000380310.2 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr11_+_66742742 | 0.07 |
ENST00000308963.4
|
C11orf86
|
chromosome 11 open reading frame 86 |
| chr12_-_52715179 | 0.07 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr5_+_140227357 | 0.07 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr9_+_34653861 | 0.07 |
ENST00000556792.1
ENST00000318041.9 ENST00000378817.4 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr12_+_48147699 | 0.07 |
ENST00000548498.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr14_+_24779340 | 0.07 |
ENST00000533293.1
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr3_+_187420101 | 0.07 |
ENST00000449623.1
ENST00000437407.1 |
RP11-211G3.3
|
Uncharacterized protein |
| chr11_+_36589547 | 0.07 |
ENST00000299440.5
|
RAG1
|
recombination activating gene 1 |
| chr12_-_58159361 | 0.07 |
ENST00000546567.1
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr11_-_66675371 | 0.07 |
ENST00000393955.2
|
PC
|
pyruvate carboxylase |
| chr16_-_67194201 | 0.07 |
ENST00000345057.4
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr6_-_32812420 | 0.07 |
ENST00000374881.2
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr17_-_8027402 | 0.07 |
ENST00000541682.2
ENST00000317814.4 ENST00000577735.1 |
HES7
|
hes family bHLH transcription factor 7 |
| chr7_-_34978980 | 0.06 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr16_-_89785777 | 0.06 |
ENST00000561976.1
|
VPS9D1
|
VPS9 domain containing 1 |
| chr6_+_26204825 | 0.06 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chrX_+_153060090 | 0.06 |
ENST00000370086.3
ENST00000370085.3 |
SSR4
|
signal sequence receptor, delta |
| chr1_-_209824643 | 0.06 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr17_-_1552282 | 0.06 |
ENST00000574810.1
|
RILP
|
Rab interacting lysosomal protein |
| chr14_+_103573853 | 0.06 |
ENST00000560304.1
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr1_-_204116078 | 0.06 |
ENST00000367198.2
ENST00000452983.1 |
ETNK2
|
ethanolamine kinase 2 |
| chr11_+_844406 | 0.06 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr9_-_136344197 | 0.06 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr16_+_23765948 | 0.06 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr17_-_61512545 | 0.06 |
ENST00000585153.1
|
CYB561
|
cytochrome b561 |
| chr19_-_33182616 | 0.06 |
ENST00000592431.1
|
CTD-2538C1.2
|
CTD-2538C1.2 |
| chr19_-_1885432 | 0.06 |
ENST00000250974.9
|
ABHD17A
|
abhydrolase domain containing 17A |
| chr2_+_95963052 | 0.05 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chr19_-_50381606 | 0.05 |
ENST00000391830.1
|
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr12_+_51632638 | 0.05 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr1_-_155006224 | 0.05 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chrX_+_153059608 | 0.05 |
ENST00000370087.1
|
SSR4
|
signal sequence receptor, delta |
| chr17_+_77018896 | 0.05 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chrX_+_70316005 | 0.05 |
ENST00000374259.3
|
FOXO4
|
forkhead box O4 |
| chr20_-_43936937 | 0.05 |
ENST00000342716.4
ENST00000353917.5 ENST00000360607.6 ENST00000372751.4 |
MATN4
|
matrilin 4 |
| chr3_-_120400960 | 0.05 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr14_-_107283278 | 0.05 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr19_-_45996465 | 0.05 |
ENST00000430715.2
|
RTN2
|
reticulon 2 |
| chr15_-_90294523 | 0.05 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr11_+_60691924 | 0.05 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr9_-_96717654 | 0.05 |
ENST00000253968.6
|
BARX1
|
BARX homeobox 1 |
| chr5_-_132166579 | 0.05 |
ENST00000378679.3
|
SHROOM1
|
shroom family member 1 |
| chr7_+_73245193 | 0.05 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr12_+_58166726 | 0.05 |
ENST00000546504.1
|
RP11-571M6.15
|
Uncharacterized protein |
| chr20_+_3190006 | 0.05 |
ENST00000380113.3
ENST00000455664.2 ENST00000399838.3 |
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr5_-_132166303 | 0.05 |
ENST00000440118.1
|
SHROOM1
|
shroom family member 1 |
| chr1_-_36789755 | 0.05 |
ENST00000270824.1
|
EVA1B
|
eva-1 homolog B (C. elegans) |
| chr1_-_1310530 | 0.05 |
ENST00000338370.3
ENST00000321751.5 ENST00000378853.3 |
AURKAIP1
|
aurora kinase A interacting protein 1 |
| chr17_+_67498396 | 0.05 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr15_-_31523036 | 0.05 |
ENST00000559094.1
ENST00000558388.2 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr17_+_36508826 | 0.05 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr9_-_136344237 | 0.05 |
ENST00000432868.1
ENST00000371899.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr6_-_32811771 | 0.05 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr1_-_2345236 | 0.05 |
ENST00000508384.1
|
PEX10
|
peroxisomal biogenesis factor 10 |
| chr7_-_150038704 | 0.05 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr11_+_62379194 | 0.05 |
ENST00000525801.1
ENST00000534093.1 |
ROM1
|
retinal outer segment membrane protein 1 |
| chr22_+_21996549 | 0.05 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr12_+_10331605 | 0.05 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr2_+_145780739 | 0.05 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr20_+_58571419 | 0.05 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr17_+_7452336 | 0.05 |
ENST00000293826.4
|
TNFSF12-TNFSF13
|
TNFSF12-TNFSF13 readthrough |
| chrX_+_48916497 | 0.05 |
ENST00000496529.2
ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120
|
coiled-coil domain containing 120 |
| chr19_+_10362882 | 0.05 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr19_+_54496132 | 0.05 |
ENST00000346968.2
|
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr9_-_92020841 | 0.05 |
ENST00000433650.1
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr6_-_44225231 | 0.05 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr17_+_14277419 | 0.05 |
ENST00000436469.1
|
AC022816.2
|
AC022816.2 |
| chr2_-_73869508 | 0.05 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr17_-_40288449 | 0.04 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr2_-_220034712 | 0.04 |
ENST00000409370.2
ENST00000430764.1 ENST00000409878.3 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr7_+_100271446 | 0.04 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr16_-_31076332 | 0.04 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr14_-_23446003 | 0.04 |
ENST00000553911.1
|
AJUBA
|
ajuba LIM protein |
| chr11_-_117698787 | 0.04 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr16_+_29911864 | 0.04 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr4_+_175204865 | 0.04 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr19_+_56652643 | 0.04 |
ENST00000586123.1
|
ZNF444
|
zinc finger protein 444 |
| chr11_-_117688216 | 0.04 |
ENST00000525836.1
|
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr16_-_85784634 | 0.04 |
ENST00000284245.4
ENST00000602914.1 |
C16orf74
|
chromosome 16 open reading frame 74 |
| chr20_+_327668 | 0.04 |
ENST00000382291.3
ENST00000609504.1 ENST00000382285.2 |
NRSN2
|
neurensin 2 |
| chr7_-_130598059 | 0.04 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr19_-_10613862 | 0.04 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr17_+_77021702 | 0.04 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr8_+_107282368 | 0.04 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr19_+_39904168 | 0.04 |
ENST00000438123.1
ENST00000409797.2 ENST00000451354.2 |
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr17_+_77019030 | 0.04 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr18_-_64271363 | 0.04 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr1_-_235098861 | 0.04 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr12_+_49740700 | 0.04 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr6_+_100054606 | 0.04 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr19_-_38806540 | 0.04 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr3_-_47950745 | 0.04 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr11_-_73687997 | 0.04 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr20_-_33460621 | 0.04 |
ENST00000427420.1
ENST00000336431.5 |
GGT7
|
gamma-glutamyltransferase 7 |
| chr8_-_21999447 | 0.04 |
ENST00000306306.3
ENST00000521744.1 |
REEP4
|
receptor accessory protein 4 |
| chrX_+_153639856 | 0.04 |
ENST00000426834.1
ENST00000369790.4 ENST00000454722.1 ENST00000350743.4 ENST00000299328.5 ENST00000351413.4 |
TAZ
|
tafazzin |
| chrX_-_48755030 | 0.04 |
ENST00000490755.2
ENST00000465150.2 ENST00000495490.2 |
TIMM17B
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr19_+_12944722 | 0.04 |
ENST00000591495.1
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr1_+_161185032 | 0.04 |
ENST00000367992.3
ENST00000289902.1 |
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr20_+_57464200 | 0.04 |
ENST00000604005.1
|
GNAS
|
GNAS complex locus |
| chr7_-_37024665 | 0.04 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr12_-_111358372 | 0.04 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr17_+_7341586 | 0.04 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr15_-_75248954 | 0.04 |
ENST00000499788.2
|
RPP25
|
ribonuclease P/MRP 25kDa subunit |
| chr6_+_43140095 | 0.04 |
ENST00000457278.2
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr12_-_7245018 | 0.04 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr1_-_153949751 | 0.04 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr15_-_70390213 | 0.04 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr6_+_31926857 | 0.04 |
ENST00000375394.2
ENST00000544581.1 |
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr17_+_42427826 | 0.04 |
ENST00000586443.1
|
GRN
|
granulin |
| chr14_-_23446021 | 0.04 |
ENST00000553592.1
|
AJUBA
|
ajuba LIM protein |
| chr19_-_45735138 | 0.04 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr9_+_35605274 | 0.04 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr17_+_73606766 | 0.04 |
ENST00000578462.1
|
MYO15B
|
myosin XVB pseudogene |
| chr19_-_6501778 | 0.04 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chr1_-_153950116 | 0.04 |
ENST00000368589.1
|
JTB
|
jumping translocation breakpoint |
| chr2_+_65663812 | 0.04 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr8_-_121824374 | 0.04 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr1_+_161195781 | 0.03 |
ENST00000367988.3
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr5_-_138730817 | 0.03 |
ENST00000434752.2
|
PROB1
|
proline-rich basic protein 1 |
| chr19_-_19314162 | 0.03 |
ENST00000420605.3
ENST00000544883.1 ENST00000538165.2 ENST00000331552.7 |
NR2C2AP
|
nuclear receptor 2C2-associated protein |
| chr16_+_30751953 | 0.03 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr11_-_1783633 | 0.03 |
ENST00000367196.3
|
CTSD
|
cathepsin D |
| chr16_+_57139933 | 0.03 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr2_-_158182322 | 0.03 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr6_-_31514333 | 0.03 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr7_-_105221898 | 0.03 |
ENST00000486180.1
ENST00000485614.1 ENST00000480514.1 |
EFCAB10
|
EF-hand calcium binding domain 10 |
| chr9_-_136203235 | 0.03 |
ENST00000372022.4
|
SURF6
|
surfeit 6 |
| chr1_-_11120057 | 0.03 |
ENST00000376957.2
|
SRM
|
spermidine synthase |
| chr1_+_155006300 | 0.03 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr17_-_43502987 | 0.03 |
ENST00000376922.2
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chrX_+_49091920 | 0.03 |
ENST00000376227.3
|
CCDC22
|
coiled-coil domain containing 22 |
| chr1_+_149754227 | 0.03 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr11_-_66725837 | 0.03 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr17_+_10048831 | 0.03 |
ENST00000581040.1
|
AC000003.2
|
CDNA FLJ25865 fis, clone CBR01927 |
| chr7_+_6522922 | 0.03 |
ENST00000601673.1
|
FLJ20306
|
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
| chr9_-_33402506 | 0.03 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr5_+_150157444 | 0.03 |
ENST00000526627.1
|
SMIM3
|
small integral membrane protein 3 |
| chr17_+_14204389 | 0.03 |
ENST00000360954.2
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
| chr2_-_158182105 | 0.03 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr6_-_31514516 | 0.03 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr2_-_31637560 | 0.03 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr2_-_96926313 | 0.03 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr16_+_28986085 | 0.03 |
ENST00000565975.1
ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr4_+_15341442 | 0.03 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr16_+_68344981 | 0.03 |
ENST00000441236.1
ENST00000348497.4 ENST00000339507.5 |
PRMT7
|
protein arginine methyltransferase 7 |
| chr13_-_114107839 | 0.03 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr12_-_2944184 | 0.03 |
ENST00000337508.4
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chr10_-_75193308 | 0.03 |
ENST00000299432.2
|
MSS51
|
MSS51 mitochondrial translational activator |
| chr12_+_6644443 | 0.03 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_153917700 | 0.03 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr8_+_97506033 | 0.03 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr7_+_100271355 | 0.03 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr5_+_137673200 | 0.03 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr17_+_46189311 | 0.03 |
ENST00000582481.1
|
SNX11
|
sorting nexin 11 |
| chr12_-_57630873 | 0.03 |
ENST00000556732.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr19_-_36233332 | 0.03 |
ENST00000592537.1
ENST00000246532.1 ENST00000344990.3 ENST00000588992.1 |
IGFLR1
|
IGF-like family receptor 1 |
| chrX_+_55026763 | 0.03 |
ENST00000374987.3
|
APEX2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr1_+_180875711 | 0.03 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr11_-_117698765 | 0.03 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr11_+_76777979 | 0.03 |
ENST00000531028.1
ENST00000278559.3 ENST00000527066.1 ENST00000529629.1 |
CAPN5
|
calpain 5 |
| chr19_-_5680499 | 0.03 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr22_+_41763274 | 0.03 |
ENST00000406644.3
|
TEF
|
thyrotrophic embryonic factor |
| chr11_-_61584233 | 0.03 |
ENST00000491310.1
|
FADS1
|
fatty acid desaturase 1 |
| chr5_-_135290705 | 0.03 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr4_-_103749205 | 0.03 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr19_-_5680891 | 0.03 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr6_+_35310312 | 0.03 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr19_+_56652686 | 0.03 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr5_-_78281623 | 0.03 |
ENST00000521117.1
|
ARSB
|
arylsulfatase B |
| chr2_+_130737223 | 0.03 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr16_+_67571351 | 0.03 |
ENST00000428437.2
ENST00000569253.1 |
FAM65A
|
family with sequence similarity 65, member A |
| chr9_+_131174024 | 0.03 |
ENST00000420034.1
ENST00000372842.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr11_+_33061336 | 0.03 |
ENST00000602733.1
|
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr13_-_80915059 | 0.03 |
ENST00000377104.3
|
SPRY2
|
sprouty homolog 2 (Drosophila) |
| chr6_+_30585486 | 0.03 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr19_-_50380536 | 0.03 |
ENST00000391832.3
ENST00000391834.2 ENST00000344175.5 |
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr19_+_17448348 | 0.03 |
ENST00000324894.8
ENST00000358792.7 ENST00000600625.1 |
GTPBP3
|
GTP binding protein 3 (mitochondrial) |
| chr20_-_62203808 | 0.03 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr1_+_155100342 | 0.03 |
ENST00000368406.2
|
EFNA1
|
ephrin-A1 |
| chr17_-_77924627 | 0.03 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr17_-_3571934 | 0.03 |
ENST00000225525.3
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr19_-_5680231 | 0.03 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
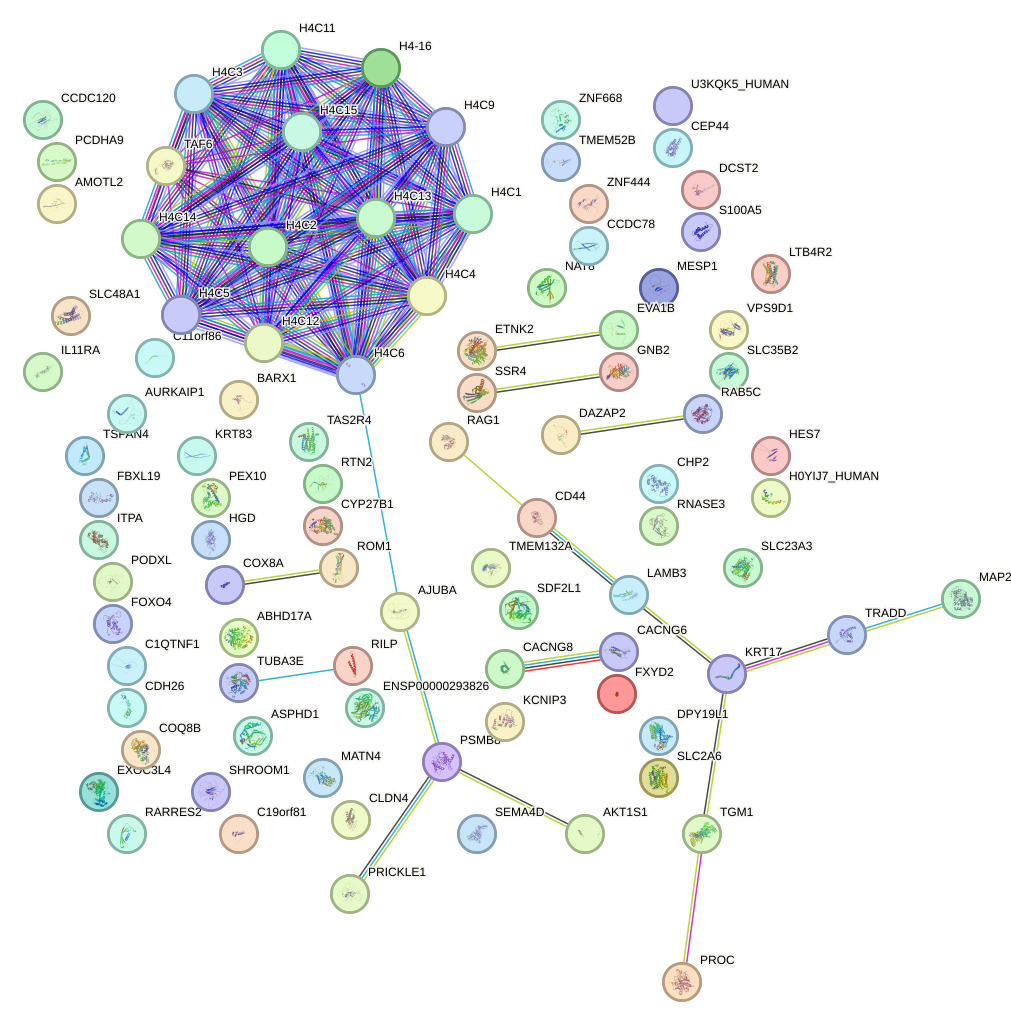
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.0 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.1 | GO:0100012 | negative regulation of mesodermal cell fate specification(GO:0042662) cardiac cell fate determination(GO:0060913) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.0 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 0.0 | GO:0061760 | antifungal humoral response(GO:0019732) antifungal innate immune response(GO:0061760) |
| 0.0 | 0.0 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0060532 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0004974 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0004766 | spermidine synthase activity(GO:0004766) |