Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CUUUGGU
Z-value: 0.62
miRNA associated with seed CUUUGGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-9-5p
|
MIMAT0000441 |
Activity profile of CUUUGGU motif
Sorted Z-values of CUUUGGU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_115913222 | 0.15 |
ENST00000259392.3
|
SLC31A2
|
solute carrier family 31 (copper transporter), member 2 |
| chr7_+_23636992 | 0.13 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr7_+_139026057 | 0.11 |
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae) |
| chr10_-_105110831 | 0.11 |
ENST00000337211.4
|
PCGF6
|
polycomb group ring finger 6 |
| chr1_-_197169672 | 0.10 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr1_-_100598444 | 0.10 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr14_-_39639523 | 0.09 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chrX_-_83442915 | 0.09 |
ENST00000262752.2
ENST00000543399.1 |
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr5_+_102455853 | 0.09 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr6_+_17600576 | 0.09 |
ENST00000259963.3
|
FAM8A1
|
family with sequence similarity 8, member A1 |
| chr8_+_67039131 | 0.09 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr10_-_38146510 | 0.08 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248 |
| chr3_-_71774516 | 0.08 |
ENST00000425534.3
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr14_-_34931458 | 0.08 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chr12_+_118814344 | 0.08 |
ENST00000397564.2
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr8_-_29120580 | 0.08 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr22_-_39548627 | 0.08 |
ENST00000216133.5
|
CBX7
|
chromobox homolog 7 |
| chr4_-_157892498 | 0.08 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr7_-_42951509 | 0.08 |
ENST00000438029.1
ENST00000432637.1 ENST00000447342.1 ENST00000431882.2 ENST00000350427.4 ENST00000425683.1 |
C7orf25
|
chromosome 7 open reading frame 25 |
| chr5_-_49737184 | 0.08 |
ENST00000508934.1
ENST00000303221.5 |
EMB
|
embigin |
| chr13_-_41240717 | 0.08 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr2_+_149632783 | 0.08 |
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C |
| chr1_-_46598284 | 0.08 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr1_-_169863016 | 0.08 |
ENST00000367772.4
ENST00000367771.6 |
SCYL3
|
SCY1-like 3 (S. cerevisiae) |
| chr6_+_42749759 | 0.08 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr8_-_99837856 | 0.08 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr3_+_191046810 | 0.07 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr10_-_27149792 | 0.07 |
ENST00000376140.3
ENST00000376170.4 |
ABI1
|
abl-interactor 1 |
| chr11_-_95657231 | 0.07 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr14_+_61995722 | 0.07 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr11_-_116968987 | 0.07 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr15_+_59730348 | 0.07 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr1_+_179923873 | 0.07 |
ENST00000367607.3
ENST00000491495.2 |
CEP350
|
centrosomal protein 350kDa |
| chrX_-_118986911 | 0.07 |
ENST00000276201.2
ENST00000345865.2 |
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr9_-_125693757 | 0.07 |
ENST00000373656.3
|
ZBTB26
|
zinc finger and BTB domain containing 26 |
| chr3_+_43328004 | 0.07 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr3_-_9291063 | 0.07 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr18_-_5296001 | 0.07 |
ENST00000357006.4
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr3_-_196159268 | 0.07 |
ENST00000381887.3
ENST00000535858.1 ENST00000428095.1 ENST00000296328.4 |
UBXN7
|
UBX domain protein 7 |
| chr12_+_27396901 | 0.07 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr3_+_183353356 | 0.07 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr5_-_169407744 | 0.07 |
ENST00000377365.3
|
FAM196B
|
family with sequence similarity 196, member B |
| chr10_+_75757863 | 0.07 |
ENST00000372755.3
ENST00000211998.4 ENST00000417648.2 |
VCL
|
vinculin |
| chr20_-_50419055 | 0.06 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr17_-_49198216 | 0.06 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr11_-_96076334 | 0.06 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr8_+_38088861 | 0.06 |
ENST00000397166.2
ENST00000533100.1 |
DDHD2
|
DDHD domain containing 2 |
| chr8_-_57123815 | 0.06 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr7_-_100183742 | 0.06 |
ENST00000310300.6
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr1_+_97187318 | 0.06 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr14_-_55878538 | 0.06 |
ENST00000247178.5
|
ATG14
|
autophagy related 14 |
| chr13_-_22033392 | 0.06 |
ENST00000320220.9
ENST00000415724.1 ENST00000422251.1 ENST00000382466.3 ENST00000542645.1 ENST00000400590.3 |
ZDHHC20
|
zinc finger, DHHC-type containing 20 |
| chr15_+_51200859 | 0.06 |
ENST00000261842.5
|
AP4E1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr11_-_94227029 | 0.06 |
ENST00000323977.3
ENST00000536754.1 ENST00000323929.3 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr22_-_36784035 | 0.06 |
ENST00000216181.5
|
MYH9
|
myosin, heavy chain 9, non-muscle |
| chr16_+_68771128 | 0.06 |
ENST00000261769.5
ENST00000422392.2 |
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr14_+_70078303 | 0.06 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247 |
| chr5_-_14871866 | 0.06 |
ENST00000284268.6
|
ANKH
|
ANKH inorganic pyrophosphate transport regulator |
| chr3_+_141205852 | 0.06 |
ENST00000286364.3
ENST00000452898.1 |
RASA2
|
RAS p21 protein activator 2 |
| chr2_+_97454321 | 0.06 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr2_+_48541776 | 0.06 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr10_+_65281123 | 0.06 |
ENST00000298249.4
ENST00000373758.4 |
REEP3
|
receptor accessory protein 3 |
| chr18_+_11851383 | 0.06 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr1_+_25071848 | 0.06 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr13_+_98086445 | 0.06 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr17_+_46908350 | 0.06 |
ENST00000258947.3
ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr9_+_108210279 | 0.06 |
ENST00000374716.4
ENST00000374710.3 ENST00000481272.1 ENST00000484973.1 ENST00000394926.3 ENST00000539376.1 |
FSD1L
|
fibronectin type III and SPRY domain containing 1-like |
| chr2_+_46769798 | 0.06 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr5_+_157170703 | 0.06 |
ENST00000286307.5
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chrX_+_103411189 | 0.06 |
ENST00000493442.1
|
FAM199X
|
family with sequence similarity 199, X-linked |
| chr10_+_121652204 | 0.06 |
ENST00000369075.3
ENST00000543134.1 |
SEC23IP
|
SEC23 interacting protein |
| chr2_+_26568965 | 0.06 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr2_+_203499901 | 0.06 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr12_+_68042495 | 0.06 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr4_+_1283639 | 0.06 |
ENST00000303400.4
ENST00000505177.2 ENST00000503653.1 ENST00000264750.6 ENST00000502558.1 ENST00000452175.2 ENST00000514708.1 |
MAEA
|
macrophage erythroblast attacher |
| chr18_+_9136758 | 0.06 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr13_-_100624012 | 0.05 |
ENST00000267294.4
|
ZIC5
|
Zic family member 5 |
| chr22_+_24666763 | 0.05 |
ENST00000437398.1
ENST00000421374.1 ENST00000314328.9 ENST00000541492.1 |
SPECC1L
|
sperm antigen with calponin homology and coiled-coil domains 1-like |
| chr4_-_78740511 | 0.05 |
ENST00000504123.1
ENST00000264903.4 ENST00000515441.1 |
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr3_+_171758344 | 0.05 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr11_+_119019722 | 0.05 |
ENST00000307417.3
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr12_+_111843749 | 0.05 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr6_-_82462425 | 0.05 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr2_-_39348137 | 0.05 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr7_+_139044621 | 0.05 |
ENST00000354926.4
|
C7orf55-LUC7L2
|
C7orf55-LUC7L2 readthrough |
| chr15_+_41952591 | 0.05 |
ENST00000566718.1
ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA
|
MGA, MAX dimerization protein |
| chr14_+_52118576 | 0.05 |
ENST00000395718.2
ENST00000344768.5 |
FRMD6
|
FERM domain containing 6 |
| chr3_+_105085734 | 0.05 |
ENST00000306107.5
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr17_-_62658186 | 0.05 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chrX_-_131352152 | 0.05 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr2_-_1748214 | 0.05 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr7_-_95225768 | 0.05 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr3_-_52713729 | 0.05 |
ENST00000296302.7
ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1
|
polybromo 1 |
| chr17_+_28804380 | 0.05 |
ENST00000225724.5
ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr17_+_45727204 | 0.05 |
ENST00000290158.4
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr12_+_72056773 | 0.05 |
ENST00000308086.2
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr20_+_32077880 | 0.05 |
ENST00000342704.6
ENST00000375279.2 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr2_-_101767715 | 0.05 |
ENST00000376840.4
ENST00000409318.1 |
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain) |
| chr5_+_170814803 | 0.05 |
ENST00000521672.1
ENST00000351986.6 ENST00000393820.2 ENST00000523622.1 |
NPM1
|
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| chr9_+_4490394 | 0.05 |
ENST00000262352.3
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr1_-_225840747 | 0.05 |
ENST00000366843.2
ENST00000366844.3 |
ENAH
|
enabled homolog (Drosophila) |
| chr11_+_120107344 | 0.05 |
ENST00000260264.4
|
POU2F3
|
POU class 2 homeobox 3 |
| chr1_+_26737253 | 0.05 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr18_+_52495426 | 0.05 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr12_-_6580094 | 0.05 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr7_+_155250824 | 0.05 |
ENST00000297375.4
|
EN2
|
engrailed homeobox 2 |
| chr1_-_70671216 | 0.05 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr17_+_4901199 | 0.05 |
ENST00000320785.5
ENST00000574165.1 |
KIF1C
|
kinesin family member 1C |
| chr7_-_6312206 | 0.05 |
ENST00000350796.3
|
CYTH3
|
cytohesin 3 |
| chr10_+_17686124 | 0.05 |
ENST00000377524.3
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr15_+_42787452 | 0.05 |
ENST00000249647.3
|
SNAP23
|
synaptosomal-associated protein, 23kDa |
| chr10_-_62704005 | 0.05 |
ENST00000337910.5
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr4_+_6642404 | 0.05 |
ENST00000507420.1
ENST00000382581.4 |
MRFAP1
|
Morf4 family associated protein 1 |
| chr11_+_118477144 | 0.05 |
ENST00000361417.2
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr14_-_39901618 | 0.05 |
ENST00000554932.1
ENST00000298097.7 |
FBXO33
|
F-box protein 33 |
| chr6_+_108881012 | 0.05 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr14_-_34420259 | 0.05 |
ENST00000250457.3
ENST00000547327.2 |
EGLN3
|
egl-9 family hypoxia-inducible factor 3 |
| chr1_+_193091080 | 0.05 |
ENST00000367435.3
|
CDC73
|
cell division cycle 73 |
| chr1_+_93811438 | 0.05 |
ENST00000370272.4
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2) |
| chr12_+_69004619 | 0.05 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr8_-_18871159 | 0.05 |
ENST00000327040.8
ENST00000440756.2 |
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr20_+_23342783 | 0.04 |
ENST00000544236.1
ENST00000338121.5 ENST00000542987.1 ENST00000424216.1 |
GZF1
|
GDNF-inducible zinc finger protein 1 |
| chr5_-_131826457 | 0.04 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr12_+_110437328 | 0.04 |
ENST00000261739.4
|
ANKRD13A
|
ankyrin repeat domain 13A |
| chr15_+_77712993 | 0.04 |
ENST00000336216.4
ENST00000381714.3 ENST00000558651.1 |
HMG20A
|
high mobility group 20A |
| chr8_-_66754172 | 0.04 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr6_-_136610911 | 0.04 |
ENST00000530767.1
ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1
|
BCL2-associated transcription factor 1 |
| chr13_-_23949671 | 0.04 |
ENST00000402364.1
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr19_-_460996 | 0.04 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr8_-_74791051 | 0.04 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr15_-_48937982 | 0.04 |
ENST00000316623.5
|
FBN1
|
fibrillin 1 |
| chr7_-_45960850 | 0.04 |
ENST00000381083.4
ENST00000381086.5 ENST00000275521.6 |
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr14_+_50359773 | 0.04 |
ENST00000298316.5
|
ARF6
|
ADP-ribosylation factor 6 |
| chr20_+_46130601 | 0.04 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr15_-_25684110 | 0.04 |
ENST00000232165.3
|
UBE3A
|
ubiquitin protein ligase E3A |
| chr4_+_108745711 | 0.04 |
ENST00000394684.4
|
SGMS2
|
sphingomyelin synthase 2 |
| chr1_-_173991434 | 0.04 |
ENST00000367696.2
|
RC3H1
|
ring finger and CCCH-type domains 1 |
| chr10_-_105677886 | 0.04 |
ENST00000224950.3
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr15_-_50978965 | 0.04 |
ENST00000560955.1
ENST00000313478.7 |
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr14_+_63671105 | 0.04 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr4_-_170533723 | 0.04 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr3_-_123603137 | 0.04 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr12_+_14518598 | 0.04 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chrY_+_15016725 | 0.04 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr6_-_3457256 | 0.04 |
ENST00000436008.2
|
SLC22A23
|
solute carrier family 22, member 23 |
| chr9_+_78505581 | 0.04 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr21_-_38362497 | 0.04 |
ENST00000427746.1
ENST00000336648.4 |
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr1_+_100111479 | 0.04 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chr2_-_119605253 | 0.04 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr1_+_100503643 | 0.04 |
ENST00000370152.3
|
HIAT1
|
hippocampus abundant transcript 1 |
| chr17_-_6459768 | 0.04 |
ENST00000421306.3
|
PITPNM3
|
PITPNM family member 3 |
| chr1_-_67896095 | 0.04 |
ENST00000370994.4
|
SERBP1
|
SERPINE1 mRNA binding protein 1 |
| chr5_-_16936340 | 0.04 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr5_+_82767284 | 0.04 |
ENST00000265077.3
|
VCAN
|
versican |
| chr14_-_92572894 | 0.04 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr2_+_135676381 | 0.04 |
ENST00000537343.1
ENST00000295238.6 ENST00000264157.5 |
CCNT2
|
cyclin T2 |
| chr12_+_27485823 | 0.04 |
ENST00000395901.2
ENST00000546179.1 |
ARNTL2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr1_-_26232951 | 0.04 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr13_+_93879085 | 0.04 |
ENST00000377047.4
|
GPC6
|
glypican 6 |
| chr17_+_47074758 | 0.04 |
ENST00000290341.3
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr5_-_178157700 | 0.04 |
ENST00000335815.2
|
ZNF354A
|
zinc finger protein 354A |
| chr13_-_77601282 | 0.04 |
ENST00000355619.5
|
FBXL3
|
F-box and leucine-rich repeat protein 3 |
| chr2_+_46926048 | 0.04 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr3_+_61547585 | 0.04 |
ENST00000295874.10
ENST00000474889.1 |
PTPRG
|
protein tyrosine phosphatase, receptor type, G |
| chr6_-_94129244 | 0.04 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr17_+_68165657 | 0.04 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr2_+_148778570 | 0.04 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr4_+_57774042 | 0.04 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr1_+_114472222 | 0.04 |
ENST00000369558.1
ENST00000369561.4 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr14_+_103243813 | 0.04 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr2_+_118846008 | 0.04 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr20_+_47538357 | 0.04 |
ENST00000371917.4
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr12_-_49110613 | 0.04 |
ENST00000261900.3
|
CCNT1
|
cyclin T1 |
| chr12_+_65004292 | 0.04 |
ENST00000542104.1
ENST00000336061.2 |
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr19_+_16222439 | 0.04 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr1_+_206858232 | 0.04 |
ENST00000294981.4
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr2_-_160472952 | 0.04 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr4_+_186125391 | 0.04 |
ENST00000504273.1
|
SNX25
|
sorting nexin 25 |
| chr15_+_59279851 | 0.04 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr2_-_102003987 | 0.04 |
ENST00000324768.5
|
CREG2
|
cellular repressor of E1A-stimulated genes 2 |
| chr12_+_113229737 | 0.04 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr10_-_15210666 | 0.04 |
ENST00000378165.4
|
NMT2
|
N-myristoyltransferase 2 |
| chr6_+_34433844 | 0.03 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr7_+_129710350 | 0.03 |
ENST00000335420.5
ENST00000463413.1 |
KLHDC10
|
kelch domain containing 10 |
| chr8_-_103425047 | 0.03 |
ENST00000520539.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr12_-_10875831 | 0.03 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr1_+_231664390 | 0.03 |
ENST00000366639.4
ENST00000413309.2 |
TSNAX
|
translin-associated factor X |
| chr1_-_235667716 | 0.03 |
ENST00000313984.3
ENST00000366600.3 |
B3GALNT2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr16_-_84150492 | 0.03 |
ENST00000343411.3
|
MBTPS1
|
membrane-bound transcription factor peptidase, site 1 |
| chr12_-_89918522 | 0.03 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr1_+_113615794 | 0.03 |
ENST00000361127.5
|
LRIG2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr15_+_101142722 | 0.03 |
ENST00000332783.7
ENST00000558747.1 ENST00000343276.4 |
ASB7
|
ankyrin repeat and SOCS box containing 7 |
| chr14_-_90085458 | 0.03 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr5_-_108745689 | 0.03 |
ENST00000361189.2
|
PJA2
|
praja ring finger 2, E3 ubiquitin protein ligase |
| chr6_-_37665751 | 0.03 |
ENST00000297153.7
ENST00000434837.3 |
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr2_+_121103706 | 0.03 |
ENST00000295228.3
|
INHBB
|
inhibin, beta B |
| chr17_+_1958388 | 0.03 |
ENST00000399849.3
|
HIC1
|
hypermethylated in cancer 1 |
| chr17_+_12569306 | 0.03 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr14_-_78083112 | 0.03 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr9_+_4662282 | 0.03 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
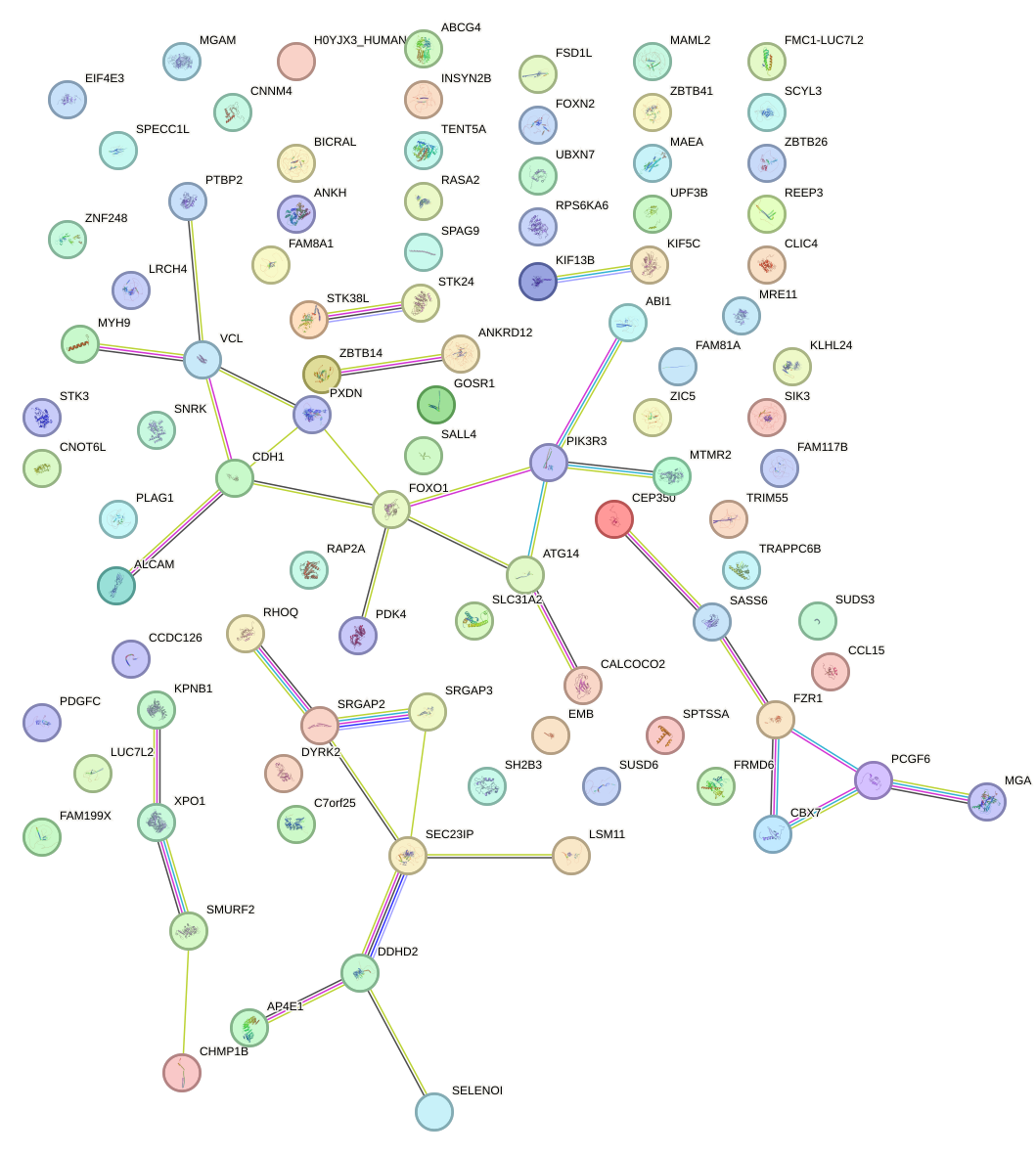
Network of associatons between targets according to the STRING database.
First level regulatory network of CUUUGGU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.0 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.0 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.0 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.0 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.0 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.0 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 0.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.0 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |