Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CUACAGU
Z-value: 0.07
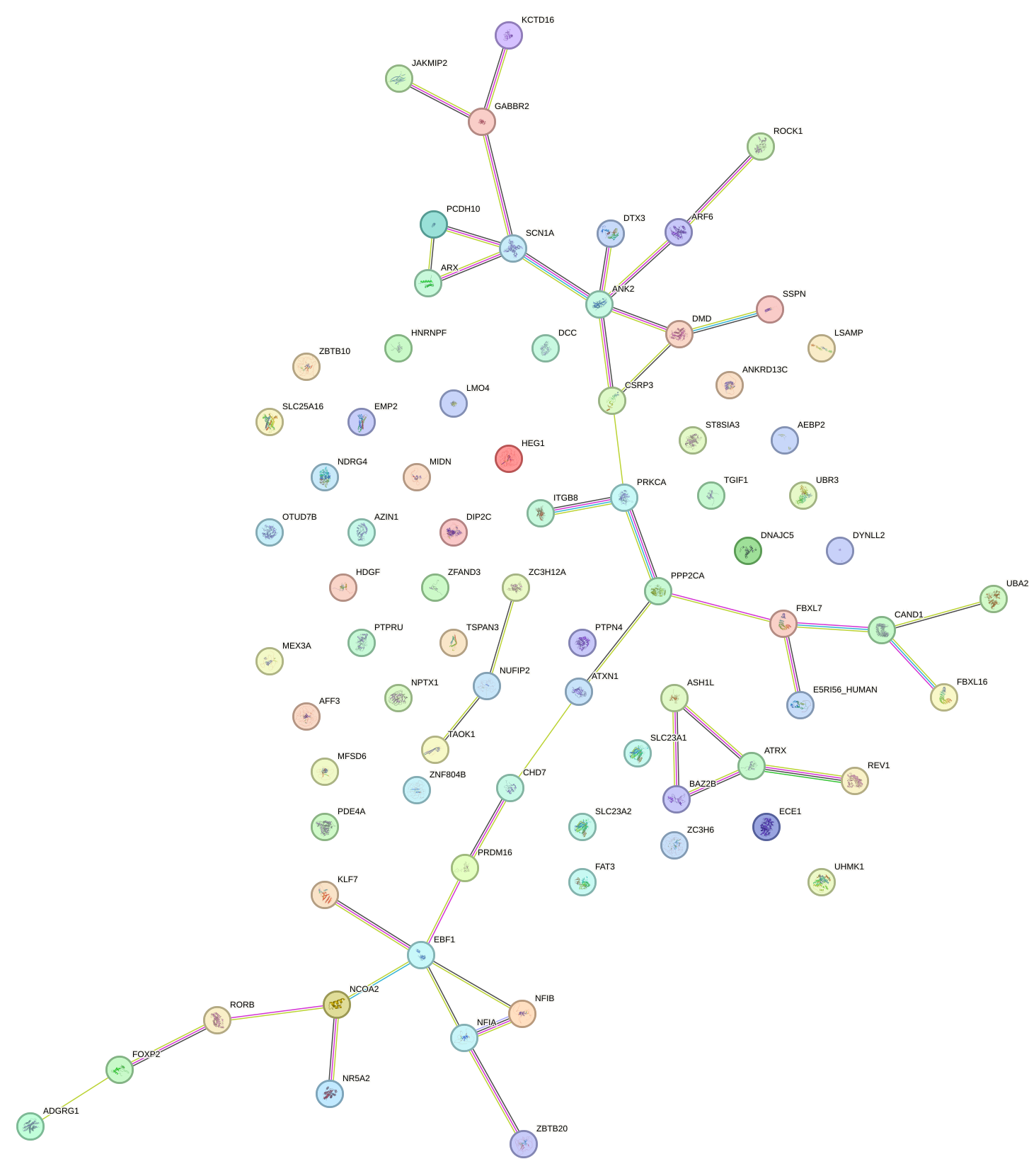
miRNA associated with seed CUACAGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-139-5p
|
MIMAT0000250 |
Activity profile of CUACAGU motif
Sorted Z-values of CUACAGU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_114790179 | 0.01 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr12_+_19592602 | 0.01 |
ENST00000398864.3
ENST00000266508.9 |
AEBP2
|
AE binding protein 2 |
| chr5_-_158526756 | 0.01 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr2_-_160472952 | 0.01 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr1_+_61547894 | 0.01 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr16_+_58497567 | 0.01 |
ENST00000258187.5
|
NDRG4
|
NDRG family member 4 |
| chrX_-_25034065 | 0.01 |
ENST00000379044.4
|
ARX
|
aristaless related homeobox |
| chr1_-_245027833 | 0.01 |
ENST00000444376.2
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr5_-_133561752 | 0.01 |
ENST00000519718.1
ENST00000481195.1 |
CTD-2410N18.5
PPP2CA
|
S-phase kinase-associated protein 1 protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr17_-_27621125 | 0.00 |
ENST00000579665.1
ENST00000225388.4 |
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr1_+_87794150 | 0.00 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr1_-_156051789 | 0.00 |
ENST00000532414.2
|
MEX3A
|
mex-3 RNA binding family member A |
| chr1_+_29563011 | 0.00 |
ENST00000345512.3
ENST00000373779.3 ENST00000356870.3 ENST00000323874.8 ENST00000428026.2 ENST00000460170.2 |
PTPRU
|
protein tyrosine phosphatase, receptor type, U |
| chr9_-_101471479 | 0.00 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr6_-_16761678 | 0.00 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr20_+_62526467 | 0.00 |
ENST00000369911.2
ENST00000360864.4 |
DNAJC5
|
DnaJ (Hsp40) homolog, subfamily C, member 5 |
| chr17_+_27717415 | 0.00 |
ENST00000583121.1
ENST00000261716.3 |
TAOK1
|
TAO kinase 1 |
| chr2_+_113033164 | 0.00 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr17_-_78450398 | 0.00 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr19_+_1249869 | 0.00 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr1_-_21671968 | 0.00 |
ENST00000415912.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr16_-_10674528 | 0.00 |
ENST00000359543.3
|
EMP2
|
epithelial membrane protein 2 |
| chr1_-_156721502 | 0.00 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr1_-_149982624 | 0.00 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr1_+_2985760 | 0.00 |
ENST00000378391.2
ENST00000514189.1 ENST00000270722.5 |
PRDM16
|
PR domain containing 16 |
| chr2_+_191273052 | 0.00 |
ENST00000417958.1
ENST00000432036.1 ENST00000392328.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr4_+_113970772 | 0.00 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr2_-_166930131 | 0.00 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr19_+_10527449 | 0.00 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chrX_-_33146477 | 0.00 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr5_+_143584814 | 0.00 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr16_-_755819 | 0.00 |
ENST00000397621.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr9_-_14314066 | 0.00 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B |
| chr14_+_50359773 | 0.00 |
ENST00000298316.5
|
ARF6
|
ADP-ribosylation factor 6 |
| chr5_+_15500280 | 0.00 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr18_-_18691739 | 0.00 |
ENST00000399799.2
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr1_+_37940153 | 0.00 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr1_-_155532484 | 0.00 |
ENST00000368346.3
ENST00000548830.1 |
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chrX_-_77041685 | 0.00 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr8_+_81397876 | 0.00 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr12_+_57998595 | 0.00 |
ENST00000337737.3
ENST00000548198.1 ENST00000551632.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr3_-_124774802 | 0.00 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr12_+_26348429 | 0.00 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr20_-_4982132 | 0.00 |
ENST00000338244.1
ENST00000424750.2 |
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr8_+_61591337 | 0.00 |
ENST00000423902.2
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr1_-_70820357 | 0.00 |
ENST00000370944.4
ENST00000262346.6 |
ANKRD13C
|
ankyrin repeat domain 13C |
| chr10_-_43903217 | 0.00 |
ENST00000357065.4
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr2_-_208030647 | 0.00 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_+_114472222 | 0.00 |
ENST00000369558.1
ENST00000369561.4 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr16_+_57662419 | 0.00 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr15_-_77363513 | 0.00 |
ENST00000267970.4
|
TSPAN3
|
tetraspanin 3 |
| chr4_+_134070439 | 0.00 |
ENST00000264360.5
|
PCDH10
|
protocadherin 10 |
| chr18_+_55018044 | 0.00 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr2_+_181845298 | 0.00 |
ENST00000410062.4
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr4_-_114682936 | 0.00 |
ENST00000454265.2
ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr9_+_77112244 | 0.00 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr2_+_170683942 | 0.00 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr10_+_82213904 | 0.00 |
ENST00000429989.3
|
TSPAN14
|
tetraspanin 14 |
| chr18_+_3451646 | 0.00 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr17_+_64298944 | 0.00 |
ENST00000413366.3
|
PRKCA
|
protein kinase C, alpha |
| chr8_-_103876965 | 0.00 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr1_+_199996702 | 0.00 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr1_+_162467595 | 0.00 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr5_-_147162078 | 0.00 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr7_+_20370746 | 0.00 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr11_+_92085262 | 0.00 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr8_-_71316021 | 0.00 |
ENST00000452400.2
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr7_+_88388628 | 0.00 |
ENST00000333190.4
|
ZNF804B
|
zinc finger protein 804B |
| chr12_+_67663056 | 0.00 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr10_-_735553 | 0.00 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr2_+_120517174 | 0.00 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr7_+_114055052 | 0.00 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr10_-_70287231 | 0.00 |
ENST00000609923.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr18_+_49866496 | 0.00 |
ENST00000442544.2
|
DCC
|
deleted in colorectal carcinoma |
| chr2_-_100721178 | 0.00 |
ENST00000409236.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr17_+_56160768 | 0.00 |
ENST00000579991.2
|
DYNLL2
|
dynein, light chain, LC8-type 2 |
| chr3_-_116164306 | 0.00 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr6_+_37787262 | 0.00 |
ENST00000287218.4
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr2_-_100106419 | 0.00 |
ENST00000393445.3
ENST00000258428.3 |
REV1
|
REV1, polymerase (DNA directed) |