Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
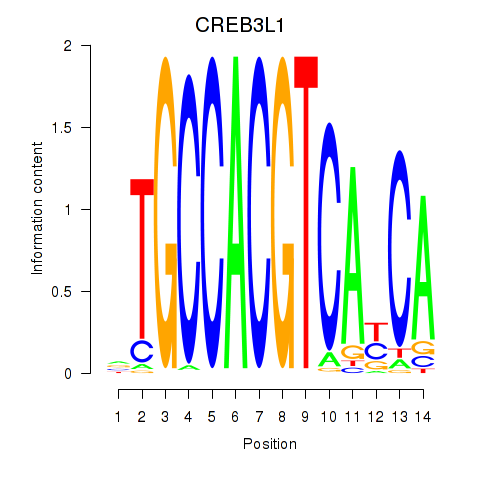
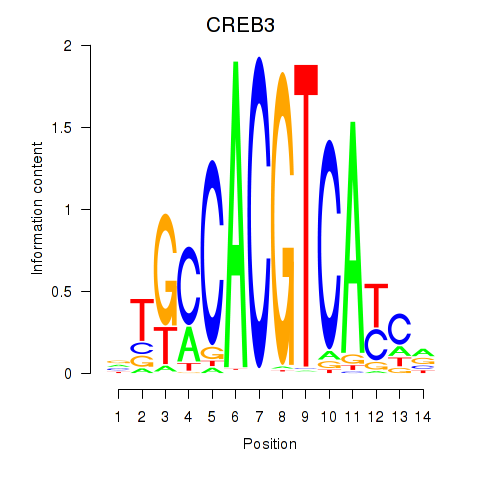
Results for CREB3L1_CREB3
Z-value: 0.41
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | cAMP responsive element binding protein 3 like 1 |
|
CREB3
|
ENSG00000107175.6 | cAMP responsive element binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L1 | hg19_v2_chr11_+_46299199_46299233 | 0.99 | 1.1e-02 | Click! |
| CREB3 | hg19_v2_chr9_+_35732312_35732332 | 0.10 | 9.0e-01 | Click! |
Activity profile of CREB3L1_CREB3 motif
Sorted Z-values of CREB3L1_CREB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_6522922 | 0.23 |
ENST00000601673.1
|
FLJ20306
|
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
| chr7_+_72848092 | 0.21 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr5_+_94890840 | 0.21 |
ENST00000504763.1
|
ARSK
|
arylsulfatase family, member K |
| chr3_-_107941209 | 0.20 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr22_-_28392227 | 0.16 |
ENST00000431039.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr17_-_191188 | 0.16 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr7_-_73184588 | 0.16 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr17_-_7137857 | 0.15 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr19_-_48894104 | 0.14 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr1_-_156399184 | 0.14 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr19_+_50353944 | 0.14 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chr17_-_7137582 | 0.11 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr19_+_50354462 | 0.11 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr7_+_140396946 | 0.10 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr3_-_178789220 | 0.10 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr3_-_119182453 | 0.10 |
ENST00000491685.1
ENST00000461654.1 |
TMEM39A
|
transmembrane protein 39A |
| chr1_-_11865982 | 0.10 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr1_+_92414952 | 0.10 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr6_+_26204825 | 0.10 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr1_+_38273818 | 0.10 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr11_-_36310958 | 0.10 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr19_+_50354393 | 0.10 |
ENST00000391842.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr19_+_50354430 | 0.09 |
ENST00000599732.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr6_-_26285737 | 0.09 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chrX_+_153059608 | 0.09 |
ENST00000370087.1
|
SSR4
|
signal sequence receptor, delta |
| chr1_+_38273988 | 0.09 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr7_+_116593953 | 0.09 |
ENST00000397750.3
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chrX_-_7895755 | 0.09 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr11_-_67276100 | 0.09 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr19_-_52097613 | 0.09 |
ENST00000301439.3
|
AC018755.1
|
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
| chr19_+_10527449 | 0.08 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chrX_+_153060090 | 0.08 |
ENST00000370086.3
ENST00000370085.3 |
SSR4
|
signal sequence receptor, delta |
| chr10_+_129785574 | 0.08 |
ENST00000430713.2
ENST00000471218.1 |
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr16_+_56642041 | 0.08 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr16_+_57220049 | 0.08 |
ENST00000562439.1
|
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr2_-_47572105 | 0.08 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr1_+_228645796 | 0.08 |
ENST00000369160.2
|
HIST3H2BB
|
histone cluster 3, H2bb |
| chr9_-_130477912 | 0.08 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr9_-_101984184 | 0.07 |
ENST00000476832.1
|
ALG2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr5_-_176730733 | 0.07 |
ENST00000504395.1
|
RAB24
|
RAB24, member RAS oncogene family |
| chr2_+_27255806 | 0.07 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr19_+_49122548 | 0.07 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr17_-_61776522 | 0.07 |
ENST00000582055.1
|
LIMD2
|
LIM domain containing 2 |
| chr1_+_145576007 | 0.07 |
ENST00000369298.1
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr3_-_156272872 | 0.07 |
ENST00000476217.1
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr3_-_132378897 | 0.07 |
ENST00000545291.1
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11 |
| chr16_-_66959429 | 0.07 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr1_+_228327943 | 0.07 |
ENST00000366726.1
ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1
|
guanylate kinase 1 |
| chr19_-_18391708 | 0.07 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr3_-_49395892 | 0.07 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chr8_+_22423168 | 0.07 |
ENST00000518912.1
ENST00000428103.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr3_-_49395705 | 0.07 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr19_-_41256207 | 0.06 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr15_-_80695917 | 0.06 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr17_+_40688190 | 0.06 |
ENST00000225927.2
|
NAGLU
|
N-acetylglucosaminidase, alpha |
| chr15_-_75249793 | 0.06 |
ENST00000322177.5
|
RPP25
|
ribonuclease P/MRP 25kDa subunit |
| chr20_+_55926625 | 0.06 |
ENST00000452119.1
|
RAE1
|
ribonucleic acid export 1 |
| chr18_-_47017956 | 0.06 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr11_-_67141640 | 0.06 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr11_-_64889649 | 0.06 |
ENST00000434372.2
|
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr14_+_103995546 | 0.06 |
ENST00000299202.4
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae) |
| chr2_-_47572207 | 0.06 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr19_-_49015050 | 0.06 |
ENST00000600059.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr11_-_118972575 | 0.06 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr1_+_41249539 | 0.06 |
ENST00000347132.5
ENST00000509682.2 |
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chr3_+_50284321 | 0.06 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr10_+_35484793 | 0.06 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr11_+_3819049 | 0.06 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr7_+_100860949 | 0.06 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr12_-_120638902 | 0.06 |
ENST00000551150.1
ENST00000313104.5 ENST00000547191.1 ENST00000546989.1 ENST00000392514.4 ENST00000228306.4 ENST00000550856.1 |
RPLP0
|
ribosomal protein, large, P0 |
| chr1_+_145575980 | 0.06 |
ENST00000393045.2
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr14_+_103995503 | 0.06 |
ENST00000389749.4
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae) |
| chr2_-_202645612 | 0.06 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chrX_+_153237740 | 0.06 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chrX_+_47441712 | 0.06 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr15_-_72668805 | 0.06 |
ENST00000268097.5
|
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr19_-_42759300 | 0.06 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr8_-_64080945 | 0.05 |
ENST00000603538.1
|
YTHDF3-AS1
|
YTHDF3 antisense RNA 1 (head to head) |
| chr19_-_4670345 | 0.05 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr16_-_69390209 | 0.05 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr11_-_119234876 | 0.05 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr3_-_57583130 | 0.05 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr19_-_3985455 | 0.05 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr17_+_73089382 | 0.05 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr16_+_56642489 | 0.05 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr19_-_40724246 | 0.05 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr1_+_228327923 | 0.05 |
ENST00000391865.3
|
GUK1
|
guanylate kinase 1 |
| chr7_-_112758589 | 0.05 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr22_-_20850070 | 0.05 |
ENST00000440659.2
ENST00000458248.1 ENST00000443285.1 ENST00000444967.1 ENST00000451553.1 ENST00000431430.1 |
KLHL22
|
kelch-like family member 22 |
| chr8_+_22423479 | 0.05 |
ENST00000522721.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr1_-_155145721 | 0.05 |
ENST00000295682.4
|
KRTCAP2
|
keratinocyte associated protein 2 |
| chr16_+_2083265 | 0.05 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chrX_+_101854096 | 0.05 |
ENST00000246174.2
ENST00000537008.1 ENST00000541409.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr12_-_49318715 | 0.05 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr19_+_50979753 | 0.05 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr19_-_5680891 | 0.05 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr6_+_26020672 | 0.05 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr19_-_50979981 | 0.05 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr20_-_1306351 | 0.05 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr12_-_53473136 | 0.05 |
ENST00000547837.1
ENST00000301463.4 |
SPRYD3
|
SPRY domain containing 3 |
| chr1_+_151372010 | 0.05 |
ENST00000290541.6
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr15_-_52030293 | 0.05 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr17_-_61777090 | 0.05 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr6_+_116692102 | 0.05 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr3_-_118959716 | 0.05 |
ENST00000467604.1
ENST00000491906.1 ENST00000475803.1 ENST00000479150.1 ENST00000470111.1 ENST00000459820.1 |
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr20_-_48782639 | 0.05 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr19_+_41256764 | 0.05 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr19_+_39903185 | 0.05 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr20_-_1306391 | 0.05 |
ENST00000339987.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr16_-_11836595 | 0.05 |
ENST00000356957.3
ENST00000283033.5 |
TXNDC11
|
thioredoxin domain containing 11 |
| chr19_-_2944907 | 0.04 |
ENST00000314531.4
|
ZNF77
|
zinc finger protein 77 |
| chr19_-_15236470 | 0.04 |
ENST00000533747.1
ENST00000598709.1 ENST00000534378.1 |
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr15_+_28623784 | 0.04 |
ENST00000526619.2
ENST00000337838.7 ENST00000532622.2 |
GOLGA8F
|
golgin A8 family, member F |
| chr7_+_139025875 | 0.04 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr6_-_44225231 | 0.04 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr17_+_4046462 | 0.04 |
ENST00000577075.2
ENST00000575251.1 ENST00000301391.3 |
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr3_-_57583052 | 0.04 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr1_+_26758849 | 0.04 |
ENST00000533087.1
ENST00000531312.1 ENST00000525165.1 ENST00000525326.1 ENST00000525546.1 ENST00000436153.2 ENST00000530781.1 |
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr11_+_65292538 | 0.04 |
ENST00000270176.5
ENST00000525364.1 ENST00000420247.2 ENST00000533862.1 ENST00000279270.6 ENST00000524944.1 |
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr3_+_47422485 | 0.04 |
ENST00000431726.1
ENST00000456221.1 ENST00000265562.4 |
PTPN23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr12_-_96336369 | 0.04 |
ENST00000546947.1
ENST00000546386.1 |
CCDC38
|
coiled-coil domain containing 38 |
| chr20_+_55926566 | 0.04 |
ENST00000411894.1
ENST00000429339.1 |
RAE1
|
ribonucleic acid export 1 |
| chr11_+_47430133 | 0.04 |
ENST00000531974.1
ENST00000531419.1 ENST00000531865.1 ENST00000362021.4 ENST00000354884.4 |
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr8_-_144623595 | 0.04 |
ENST00000262577.5
|
ZC3H3
|
zinc finger CCCH-type containing 3 |
| chr5_+_68389807 | 0.04 |
ENST00000380860.4
ENST00000504103.1 ENST00000502979.1 |
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr11_+_67159416 | 0.04 |
ENST00000307980.2
ENST00000544620.1 |
RAD9A
|
RAD9 homolog A (S. pombe) |
| chr2_+_220042933 | 0.04 |
ENST00000430297.2
|
FAM134A
|
family with sequence similarity 134, member A |
| chr17_-_4607335 | 0.04 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr11_+_64889773 | 0.04 |
ENST00000534078.1
ENST00000526171.1 ENST00000279242.2 ENST00000531705.1 ENST00000533943.1 |
MRPL49
|
mitochondrial ribosomal protein L49 |
| chr9_-_98079965 | 0.04 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr7_+_100271355 | 0.04 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr12_+_113376249 | 0.04 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chrX_+_101854426 | 0.04 |
ENST00000536530.1
ENST00000473968.1 ENST00000604957.1 ENST00000477663.1 ENST00000479502.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr3_+_140981456 | 0.04 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr1_-_113498943 | 0.04 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr3_+_121554046 | 0.04 |
ENST00000273668.2
ENST00000451944.2 |
EAF2
|
ELL associated factor 2 |
| chr19_-_15236562 | 0.04 |
ENST00000263383.3
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr10_-_22292675 | 0.03 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr15_-_28778117 | 0.03 |
ENST00000525590.2
ENST00000329523.6 |
GOLGA8G
|
golgin A8 family, member G |
| chr22_-_20850128 | 0.03 |
ENST00000328879.4
|
KLHL22
|
kelch-like family member 22 |
| chr7_+_100271446 | 0.03 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr9_-_131709858 | 0.03 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr16_+_68057179 | 0.03 |
ENST00000567100.1
ENST00000432752.1 ENST00000569289.1 ENST00000564781.1 |
DUS2
|
dihydrouridine synthase 2 |
| chr3_+_3168692 | 0.03 |
ENST00000402675.1
ENST00000413000.1 |
TRNT1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr2_-_98280383 | 0.03 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr1_+_26758790 | 0.03 |
ENST00000427245.2
ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr7_+_99070527 | 0.03 |
ENST00000379724.3
|
ZNF789
|
zinc finger protein 789 |
| chr11_-_67141090 | 0.03 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chrX_-_153236819 | 0.03 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr19_-_55574538 | 0.03 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr18_-_46895066 | 0.03 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr18_-_47018897 | 0.03 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr3_-_156272924 | 0.03 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr18_-_47018769 | 0.03 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr17_-_7145475 | 0.03 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr5_-_114961673 | 0.03 |
ENST00000333314.3
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr2_-_114514181 | 0.03 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chrX_+_152240819 | 0.03 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr1_-_32110467 | 0.03 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr5_+_176730769 | 0.03 |
ENST00000303204.4
ENST00000503216.1 |
PRELID1
|
PRELI domain containing 1 |
| chr8_-_29208183 | 0.03 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr20_+_55926583 | 0.03 |
ENST00000395840.2
|
RAE1
|
ribonucleic acid export 1 |
| chr16_+_68056844 | 0.03 |
ENST00000565263.1
|
DUS2
|
dihydrouridine synthase 2 |
| chr3_-_4508925 | 0.03 |
ENST00000534863.1
ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1
|
sulfatase modifying factor 1 |
| chr12_-_56321397 | 0.03 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr18_-_47018869 | 0.03 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr1_-_202311088 | 0.03 |
ENST00000367274.4
|
UBE2T
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr17_+_6554971 | 0.03 |
ENST00000391428.2
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr5_-_172198190 | 0.03 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr7_+_102073966 | 0.03 |
ENST00000495936.1
ENST00000356387.2 ENST00000478730.2 ENST00000468241.1 ENST00000403646.3 |
ORAI2
|
ORAI calcium release-activated calcium modulator 2 |
| chrX_+_48660287 | 0.03 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr16_-_81040719 | 0.03 |
ENST00000219400.3
|
CMC2
|
C-x(9)-C motif containing 2 |
| chr12_+_107349606 | 0.03 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr7_-_100860851 | 0.03 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr7_+_89975979 | 0.03 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr15_-_72668185 | 0.02 |
ENST00000457859.2
ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr12_+_44229846 | 0.02 |
ENST00000551577.1
ENST00000266534.3 |
TMEM117
|
transmembrane protein 117 |
| chr7_-_91808441 | 0.02 |
ENST00000437357.1
ENST00000458448.1 |
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr14_+_101293687 | 0.02 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr15_+_40987327 | 0.02 |
ENST00000423169.2
ENST00000267868.3 ENST00000557850.1 ENST00000532743.1 ENST00000382643.3 |
RAD51
|
RAD51 recombinase |
| chr12_+_2921788 | 0.02 |
ENST00000228799.2
ENST00000419778.2 ENST00000542548.1 |
ITFG2
|
integrin alpha FG-GAP repeat containing 2 |
| chr7_+_30174426 | 0.02 |
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr4_-_54930790 | 0.02 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr14_-_35344093 | 0.02 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr20_+_23331373 | 0.02 |
ENST00000254998.2
|
NXT1
|
NTF2-like export factor 1 |
| chr2_-_43823093 | 0.02 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr13_+_49822041 | 0.02 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr15_+_72947079 | 0.02 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chr2_-_43823119 | 0.02 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr1_-_153940097 | 0.02 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr19_-_41196534 | 0.02 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr4_+_165675197 | 0.02 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr15_+_91498089 | 0.02 |
ENST00000394258.2
ENST00000555155.1 |
RCCD1
|
RCC1 domain containing 1 |
| chr10_+_129785536 | 0.02 |
ENST00000419012.2
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr1_-_113498616 | 0.02 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr7_+_30174574 | 0.02 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr19_-_46088068 | 0.02 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr7_+_150929550 | 0.02 |
ENST00000482173.1
ENST00000495645.1 ENST00000035307.2 |
CHPF2
|
chondroitin polymerizing factor 2 |
| chr2_-_220042825 | 0.02 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr16_-_2059748 | 0.02 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L1_CREB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.3 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.0 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.0 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.0 | GO:0032783 | ELL-EAF complex(GO:0032783) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.0 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |