Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
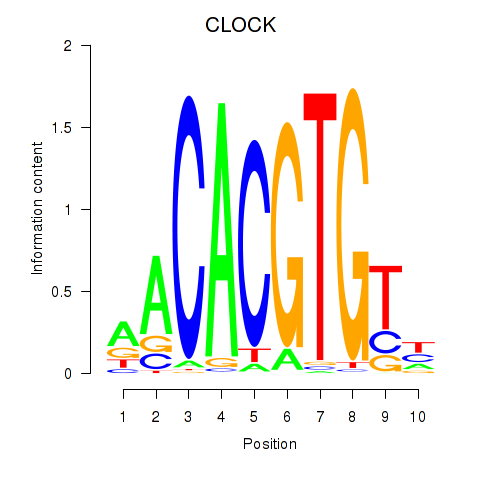
Results for CLOCK
Z-value: 0.91
Transcription factors associated with CLOCK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CLOCK
|
ENSG00000134852.10 | clock circadian regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CLOCK | hg19_v2_chr4_-_56412713_56412799 | 0.61 | 3.9e-01 | Click! |
Activity profile of CLOCK motif
Sorted Z-values of CLOCK motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_232328867 | 0.94 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr15_+_44084040 | 0.46 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr15_+_44084503 | 0.42 |
ENST00000409960.2
ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2
|
small EDRK-rich factor 2 |
| chr1_+_150254936 | 0.40 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr7_-_130597935 | 0.39 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr13_-_44735393 | 0.37 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr4_-_1107306 | 0.37 |
ENST00000433731.2
ENST00000333673.5 ENST00000382968.5 |
RNF212
|
ring finger protein 212 |
| chr9_-_100684769 | 0.37 |
ENST00000455506.1
ENST00000375117.4 |
C9orf156
|
chromosome 9 open reading frame 156 |
| chr1_-_26233423 | 0.34 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr2_+_198365095 | 0.33 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr1_-_231376836 | 0.32 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr3_-_150481218 | 0.31 |
ENST00000482706.1
|
SIAH2
|
siah E3 ubiquitin protein ligase 2 |
| chr4_+_183065793 | 0.30 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr2_-_198364581 | 0.29 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr5_+_176784837 | 0.28 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr9_-_135545380 | 0.28 |
ENST00000544003.1
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr16_+_50280020 | 0.26 |
ENST00000564965.1
|
ADCY7
|
adenylate cyclase 7 |
| chr19_-_39360561 | 0.25 |
ENST00000593809.1
ENST00000593424.1 |
RINL
|
Ras and Rab interactor-like |
| chr12_+_28343365 | 0.25 |
ENST00000545336.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_-_26232951 | 0.25 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr10_-_6019455 | 0.25 |
ENST00000530685.1
ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr3_-_42306248 | 0.25 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr5_+_43120985 | 0.24 |
ENST00000515326.1
|
ZNF131
|
zinc finger protein 131 |
| chr16_-_18937072 | 0.24 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr9_+_131445703 | 0.23 |
ENST00000454747.1
|
SET
|
SET nuclear oncogene |
| chr6_-_80657292 | 0.23 |
ENST00000369816.4
|
ELOVL4
|
ELOVL fatty acid elongase 4 |
| chr11_+_72929402 | 0.23 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr5_-_102455801 | 0.22 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr5_+_75904918 | 0.22 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr12_-_13529594 | 0.22 |
ENST00000539026.1
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr2_+_232575128 | 0.22 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr2_+_187350973 | 0.22 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr18_-_2982869 | 0.21 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr6_-_146056341 | 0.21 |
ENST00000435470.1
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr2_+_198365122 | 0.21 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr17_-_49021974 | 0.21 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr3_-_53878644 | 0.21 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr4_+_75023816 | 0.20 |
ENST00000395759.2
ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr19_+_46003056 | 0.20 |
ENST00000401593.1
ENST00000396736.2 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr17_+_73642315 | 0.20 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr22_-_42310570 | 0.20 |
ENST00000457093.1
|
SHISA8
|
shisa family member 8 |
| chr7_+_23636992 | 0.20 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr3_+_122785895 | 0.19 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr9_+_37120536 | 0.19 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr1_+_244998918 | 0.19 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr6_+_146056706 | 0.19 |
ENST00000603994.1
|
RP3-466P17.1
|
RP3-466P17.1 |
| chr12_-_123634449 | 0.19 |
ENST00000542210.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr7_-_92465868 | 0.18 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr5_+_34656450 | 0.18 |
ENST00000514527.1
|
RAI14
|
retinoic acid induced 14 |
| chr1_+_93645314 | 0.18 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr1_-_197744763 | 0.18 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr5_+_110427983 | 0.18 |
ENST00000513710.2
ENST00000505303.1 |
WDR36
|
WD repeat domain 36 |
| chr8_+_104383728 | 0.18 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr3_-_112280709 | 0.17 |
ENST00000402314.2
ENST00000283290.5 ENST00000492886.1 |
ATG3
|
autophagy related 3 |
| chr8_+_86157699 | 0.17 |
ENST00000321764.3
|
CA13
|
carbonic anhydrase XIII |
| chr4_-_75024085 | 0.17 |
ENST00000600169.1
|
AC093677.1
|
Uncharacterized protein |
| chr9_+_96793076 | 0.17 |
ENST00000375360.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr16_-_4588469 | 0.17 |
ENST00000588381.1
ENST00000563332.2 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr6_-_153304148 | 0.17 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chr6_-_114292284 | 0.17 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr11_-_3013482 | 0.17 |
ENST00000529361.1
ENST00000528968.1 ENST00000534372.1 ENST00000531291.1 ENST00000526842.1 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr14_-_91884150 | 0.17 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr16_-_81129845 | 0.16 |
ENST00000569885.1
ENST00000566566.1 |
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr8_+_38614754 | 0.16 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr9_-_100684845 | 0.16 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr2_+_173420697 | 0.16 |
ENST00000282077.3
ENST00000392571.2 ENST00000410055.1 |
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr12_-_6715808 | 0.16 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr19_-_49137790 | 0.16 |
ENST00000599385.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr5_+_102455853 | 0.16 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr4_-_83351294 | 0.16 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr1_+_38478432 | 0.16 |
ENST00000537711.1
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr9_+_37079888 | 0.16 |
ENST00000429493.1
ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1
|
RP11-220I1.1 |
| chr2_-_4703793 | 0.16 |
ENST00000421212.1
ENST00000412134.1 |
AC022311.1
|
AC022311.1 |
| chr8_+_109455845 | 0.16 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr1_-_231376867 | 0.16 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr12_+_64798095 | 0.16 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr12_-_105352080 | 0.15 |
ENST00000433540.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chrX_-_101186981 | 0.15 |
ENST00000458570.1
|
ZMAT1
|
zinc finger, matrin-type 1 |
| chr2_+_131769256 | 0.15 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr13_-_36788718 | 0.15 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr3_+_99536663 | 0.15 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr12_-_76477707 | 0.15 |
ENST00000551992.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr15_+_98503922 | 0.15 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr2_+_201171268 | 0.15 |
ENST00000423749.1
ENST00000428692.1 ENST00000457757.1 ENST00000453663.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr3_+_52720187 | 0.15 |
ENST00000474423.1
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr2_+_203130439 | 0.15 |
ENST00000264279.5
|
NOP58
|
NOP58 ribonucleoprotein |
| chr17_-_40333099 | 0.15 |
ENST00000607371.1
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr3_-_188665428 | 0.15 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr21_-_46237883 | 0.15 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr2_+_201171064 | 0.15 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr11_-_94226964 | 0.15 |
ENST00000538923.1
ENST00000540013.1 ENST00000407439.3 ENST00000393241.4 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr6_+_149539053 | 0.14 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr6_-_153304697 | 0.14 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr2_+_111878483 | 0.14 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr19_-_49828438 | 0.14 |
ENST00000454748.3
ENST00000598828.1 ENST00000335875.4 |
SLC6A16
|
solute carrier family 6, member 16 |
| chr1_+_214776516 | 0.14 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr6_-_114292449 | 0.14 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr11_+_72929319 | 0.14 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr5_+_138609782 | 0.14 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr1_+_92545862 | 0.14 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr16_+_66914264 | 0.14 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr4_+_56212505 | 0.14 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr1_+_150898733 | 0.14 |
ENST00000525956.1
|
SETDB1
|
SET domain, bifurcated 1 |
| chr5_+_138609441 | 0.14 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr17_+_76311791 | 0.14 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr21_-_16374688 | 0.14 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr6_+_131571535 | 0.14 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr4_-_76649546 | 0.14 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr8_+_107593198 | 0.14 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr11_-_94227029 | 0.13 |
ENST00000323977.3
ENST00000536754.1 ENST00000323929.3 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr19_-_47164386 | 0.13 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr7_-_50633078 | 0.13 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr2_-_232329186 | 0.13 |
ENST00000322723.4
|
NCL
|
nucleolin |
| chr11_+_34643600 | 0.13 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr8_+_6566206 | 0.13 |
ENST00000518327.1
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr14_-_91884115 | 0.13 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr5_+_34656529 | 0.13 |
ENST00000513974.1
ENST00000512629.1 |
RAI14
|
retinoic acid induced 14 |
| chr10_+_70715884 | 0.13 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr5_-_176738883 | 0.13 |
ENST00000513169.1
ENST00000423571.2 ENST00000502529.1 ENST00000427908.2 |
MXD3
|
MAX dimerization protein 3 |
| chr8_+_104383759 | 0.13 |
ENST00000415886.2
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr12_-_105352047 | 0.13 |
ENST00000432951.1
ENST00000415674.1 ENST00000424946.1 |
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr16_+_84002234 | 0.13 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr9_-_6015607 | 0.13 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr4_-_18023350 | 0.13 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chr7_+_100318423 | 0.13 |
ENST00000252723.2
|
EPO
|
erythropoietin |
| chr4_+_76649753 | 0.13 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr6_-_99873145 | 0.13 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr11_-_32457176 | 0.12 |
ENST00000332351.3
|
WT1
|
Wilms tumor 1 |
| chr20_-_5931051 | 0.12 |
ENST00000453074.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr2_-_68384603 | 0.12 |
ENST00000406245.2
ENST00000409164.1 ENST00000295121.6 |
WDR92
|
WD repeat domain 92 |
| chr11_-_3013497 | 0.12 |
ENST00000448187.1
ENST00000532325.2 ENST00000399614.2 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr15_+_36871806 | 0.12 |
ENST00000566621.1
ENST00000564586.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr15_-_49447771 | 0.12 |
ENST00000558843.1
ENST00000542928.1 ENST00000561248.1 |
COPS2
|
COP9 signalosome subunit 2 |
| chr11_+_33037401 | 0.12 |
ENST00000241051.3
|
DEPDC7
|
DEP domain containing 7 |
| chr3_+_159706537 | 0.12 |
ENST00000305579.2
ENST00000480787.1 ENST00000466512.1 |
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| chr1_+_154909803 | 0.12 |
ENST00000604546.1
|
RP11-307C12.13
|
RP11-307C12.13 |
| chr1_-_28969517 | 0.12 |
ENST00000263974.4
ENST00000373824.4 |
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr20_-_5931109 | 0.12 |
ENST00000203001.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr11_+_114310237 | 0.12 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr4_+_76439095 | 0.12 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr10_-_6019552 | 0.12 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr6_-_160210604 | 0.12 |
ENST00000420894.2
ENST00000539756.1 ENST00000544255.1 |
TCP1
|
t-complex 1 |
| chr7_-_148725544 | 0.12 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr15_-_101817492 | 0.12 |
ENST00000528346.1
ENST00000531964.1 |
VIMP
|
VCP-interacting membrane protein |
| chr7_-_91509972 | 0.12 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr11_+_9595180 | 0.12 |
ENST00000450114.2
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr10_+_73156664 | 0.12 |
ENST00000398809.4
ENST00000398842.3 ENST00000461841.3 ENST00000299366.7 |
CDH23
|
cadherin-related 23 |
| chr17_-_61850894 | 0.12 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr1_+_29213678 | 0.11 |
ENST00000347529.3
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr2_+_201171372 | 0.11 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chrX_+_24711997 | 0.11 |
ENST00000379068.3
ENST00000379059.3 |
POLA1
|
polymerase (DNA directed), alpha 1, catalytic subunit |
| chr6_+_73331918 | 0.11 |
ENST00000402622.2
ENST00000355635.3 ENST00000403813.2 ENST00000414165.2 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr1_+_29213584 | 0.11 |
ENST00000343067.4
ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr16_+_89238149 | 0.11 |
ENST00000289746.2
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr2_-_61389168 | 0.11 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr6_+_12007897 | 0.11 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr11_+_114310102 | 0.11 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr4_+_76649797 | 0.11 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr9_-_88874519 | 0.11 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chr2_-_172291273 | 0.11 |
ENST00000442778.1
ENST00000453846.1 |
METTL8
|
methyltransferase like 8 |
| chr10_-_69597828 | 0.11 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr19_+_19516561 | 0.11 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr13_-_36920615 | 0.11 |
ENST00000494062.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr17_-_40333150 | 0.11 |
ENST00000264661.3
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr4_+_111397216 | 0.11 |
ENST00000265162.5
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A) |
| chr22_-_44258280 | 0.11 |
ENST00000540422.1
|
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr13_+_98628886 | 0.11 |
ENST00000490680.1
ENST00000539640.1 ENST00000403772.3 |
IPO5
|
importin 5 |
| chr4_-_4544061 | 0.11 |
ENST00000507908.1
|
STX18
|
syntaxin 18 |
| chr5_-_176900610 | 0.11 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr17_-_17942473 | 0.11 |
ENST00000585101.1
ENST00000474627.3 ENST00000444058.1 |
ATPAF2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr2_+_187350883 | 0.11 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr11_+_3011093 | 0.11 |
ENST00000332881.2
|
AC131971.1
|
HCG1782999; PRO0943; Uncharacterized protein |
| chr17_+_40985407 | 0.11 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr5_+_102455968 | 0.11 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr5_-_93447333 | 0.10 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr11_+_114310164 | 0.10 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr17_+_21279509 | 0.10 |
ENST00000583088.1
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr1_+_1266654 | 0.10 |
ENST00000339381.5
|
TAS1R3
|
taste receptor, type 1, member 3 |
| chr2_-_74618907 | 0.10 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr11_-_1912084 | 0.10 |
ENST00000391480.1
|
C11orf89
|
chromosome 11 open reading frame 89 |
| chr16_+_1306060 | 0.10 |
ENST00000397534.2
|
TPSD1
|
tryptase delta 1 |
| chr4_+_172734548 | 0.10 |
ENST00000506823.1
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr13_-_95953589 | 0.10 |
ENST00000538287.1
ENST00000376887.4 ENST00000412704.1 ENST00000536256.1 ENST00000431522.1 |
ABCC4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr1_+_38478378 | 0.10 |
ENST00000373014.4
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr7_-_37382683 | 0.10 |
ENST00000455879.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr3_-_52719810 | 0.10 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr2_+_13677795 | 0.10 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr15_-_101835414 | 0.10 |
ENST00000254193.6
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr20_+_5931275 | 0.10 |
ENST00000378896.3
ENST00000378883.1 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr10_+_180405 | 0.10 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr12_-_54020107 | 0.10 |
ENST00000588232.1
ENST00000548446.2 ENST00000420353.2 ENST00000591397.1 ENST00000415113.1 |
ATF7
|
activating transcription factor 7 |
| chr2_-_175113088 | 0.10 |
ENST00000409546.1
ENST00000428402.2 |
OLA1
|
Obg-like ATPase 1 |
| chr1_-_78149041 | 0.10 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr13_+_27998681 | 0.10 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr3_+_52719936 | 0.10 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr9_-_98269481 | 0.10 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr8_-_103424916 | 0.10 |
ENST00000220959.4
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr16_+_53088885 | 0.10 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_+_172386517 | 0.10 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26-like 1 |
| chr19_+_1407733 | 0.10 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CLOCK
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.4 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.1 | 1.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.2 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.4 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.2 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0051808 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.3 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) brainstem development(GO:0003360) |
| 0.0 | 0.0 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.1 | GO:0044266 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0097319 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.0 | GO:0097477 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0090526 | positive regulation of glucokinase activity(GO:0033133) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.2 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.1 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.0 | 0.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.0 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.5 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.0 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |