Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
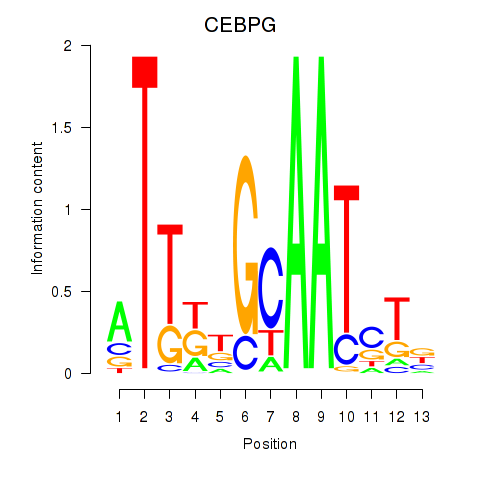
Results for CEBPG
Z-value: 0.39
Transcription factors associated with CEBPG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPG
|
ENSG00000153879.4 | CCAAT enhancer binding protein gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPG | hg19_v2_chr19_+_33865218_33865254 | 0.76 | 2.4e-01 | Click! |
Activity profile of CEBPG motif
Sorted Z-values of CEBPG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39786962 | 0.25 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr1_+_245133062 | 0.21 |
ENST00000366523.1
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr7_-_112635675 | 0.19 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr14_+_45366518 | 0.15 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr6_-_30080863 | 0.14 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr9_+_130911770 | 0.13 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr12_-_95510743 | 0.13 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr5_+_49962772 | 0.12 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_-_86353510 | 0.12 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr10_+_26986582 | 0.11 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr9_+_86237963 | 0.11 |
ENST00000277124.8
|
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr5_-_121413974 | 0.10 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr16_-_87812735 | 0.10 |
ENST00000570159.1
|
RP4-536B24.4
|
RP4-536B24.4 |
| chr12_-_13248598 | 0.09 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr20_+_54987168 | 0.09 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr1_-_114430169 | 0.09 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr8_+_97597148 | 0.09 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr10_-_14596140 | 0.09 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr19_+_37808831 | 0.09 |
ENST00000589801.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr3_-_158390282 | 0.08 |
ENST00000264265.3
|
LXN
|
latexin |
| chr12_+_27849378 | 0.08 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr6_-_30080876 | 0.08 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr19_-_6670128 | 0.08 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr14_+_45366472 | 0.08 |
ENST00000325192.3
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr20_+_54987305 | 0.08 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr6_+_36238237 | 0.08 |
ENST00000457797.1
ENST00000394571.2 |
PNPLA1
|
patatin-like phospholipase domain containing 1 |
| chr5_-_36152031 | 0.08 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr10_-_92681033 | 0.08 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr12_-_8693539 | 0.07 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr4_-_119274121 | 0.07 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr19_+_2476116 | 0.07 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr13_-_36788718 | 0.07 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr21_-_35899113 | 0.07 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr12_+_72058130 | 0.07 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr9_+_107266455 | 0.07 |
ENST00000334726.2
|
OR13F1
|
olfactory receptor, family 13, subfamily F, member 1 |
| chr14_+_96000930 | 0.06 |
ENST00000331334.4
|
GLRX5
|
glutaredoxin 5 |
| chr9_+_110045418 | 0.06 |
ENST00000419616.1
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr17_-_61850894 | 0.06 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr5_-_180552304 | 0.06 |
ENST00000329365.2
|
OR2V1
|
olfactory receptor, family 2, subfamily V, member 1 |
| chr7_-_14026063 | 0.06 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr19_-_6720686 | 0.06 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr16_-_57219926 | 0.06 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr20_-_48782639 | 0.06 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr4_-_111120334 | 0.06 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr10_+_17272608 | 0.06 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr7_-_38948774 | 0.06 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr4_+_71600144 | 0.06 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr9_+_130911723 | 0.06 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr7_+_22766766 | 0.06 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr5_-_145562147 | 0.06 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr12_-_96390108 | 0.05 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr3_+_157154578 | 0.05 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr11_-_102668879 | 0.05 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr14_-_50319758 | 0.05 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr2_-_32390801 | 0.05 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr3_-_148939598 | 0.05 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr3_-_42306248 | 0.05 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr12_-_323689 | 0.05 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr2_+_226265364 | 0.05 |
ENST00000272907.6
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr14_+_39703112 | 0.05 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr2_-_21266935 | 0.04 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chrX_-_135333722 | 0.04 |
ENST00000316077.9
|
MAP7D3
|
MAP7 domain containing 3 |
| chr16_+_57220193 | 0.04 |
ENST00000564435.1
ENST00000562959.1 ENST00000394420.4 ENST00000568505.2 ENST00000537866.1 |
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr14_-_50319482 | 0.04 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr4_+_184020398 | 0.04 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr22_-_30867973 | 0.04 |
ENST00000402286.1
ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chr6_-_30710265 | 0.04 |
ENST00000438162.1
ENST00000454845.1 |
FLOT1
|
flotillin 1 |
| chr17_-_3182268 | 0.04 |
ENST00000408891.2
|
OR3A2
|
olfactory receptor, family 3, subfamily A, member 2 |
| chr1_+_111770294 | 0.04 |
ENST00000474304.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr1_+_111770278 | 0.04 |
ENST00000369748.4
|
CHI3L2
|
chitinase 3-like 2 |
| chr12_-_96390063 | 0.04 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr5_-_179050066 | 0.04 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr19_-_36822551 | 0.04 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr3_-_148939835 | 0.04 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr16_-_3350614 | 0.04 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr17_+_7477040 | 0.04 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr6_+_74405804 | 0.04 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr12_+_32638897 | 0.04 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_165864800 | 0.04 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr4_+_71600063 | 0.03 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_-_15661474 | 0.03 |
ENST00000509314.1
ENST00000503196.1 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr11_-_124806297 | 0.03 |
ENST00000298251.4
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr6_+_74405501 | 0.03 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr12_-_54758251 | 0.03 |
ENST00000267015.3
ENST00000551809.1 |
GPR84
|
G protein-coupled receptor 84 |
| chr2_-_152146385 | 0.03 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr12_+_57522692 | 0.03 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr4_-_111119804 | 0.03 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr17_+_61851504 | 0.03 |
ENST00000359353.5
ENST00000389924.2 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr1_+_158979792 | 0.03 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_165864821 | 0.03 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr18_-_29340827 | 0.03 |
ENST00000269205.5
|
SLC25A52
|
solute carrier family 25, member 52 |
| chr4_+_71248795 | 0.03 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr1_+_158979680 | 0.03 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr12_-_69080590 | 0.03 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr5_+_49963239 | 0.03 |
ENST00000505554.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_-_175711978 | 0.02 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr2_+_149402009 | 0.02 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr16_-_57219966 | 0.02 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr17_+_61851157 | 0.02 |
ENST00000578681.1
ENST00000583590.1 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr6_+_37897735 | 0.02 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr12_-_12491608 | 0.02 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr6_+_31895467 | 0.02 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr4_-_111120132 | 0.02 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr3_+_130569429 | 0.02 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr19_+_55417499 | 0.02 |
ENST00000291890.4
ENST00000447255.1 ENST00000598576.1 ENST00000594765.1 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr11_-_72504681 | 0.02 |
ENST00000538536.1
ENST00000543304.1 ENST00000540587.1 ENST00000334805.6 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr7_-_14026123 | 0.02 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr17_-_39769005 | 0.02 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr12_+_53846612 | 0.02 |
ENST00000551104.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr3_-_4793274 | 0.02 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr1_+_93645314 | 0.02 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr20_-_60294804 | 0.02 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr20_+_361261 | 0.02 |
ENST00000217233.3
|
TRIB3
|
tribbles pseudokinase 3 |
| chr22_-_31364187 | 0.02 |
ENST00000215862.4
ENST00000397641.3 |
MORC2
|
MORC family CW-type zinc finger 2 |
| chr1_+_158979686 | 0.02 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr19_-_54327542 | 0.02 |
ENST00000391775.3
ENST00000324134.6 ENST00000535162.1 ENST00000351894.4 ENST00000354278.3 ENST00000391773.1 ENST00000345770.5 ENST00000391772.1 |
NLRP12
|
NLR family, pyrin domain containing 12 |
| chr13_+_30002741 | 0.02 |
ENST00000380808.2
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr12_+_53846594 | 0.02 |
ENST00000550192.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr17_-_29645836 | 0.01 |
ENST00000578584.1
|
CTD-2370N5.3
|
CTD-2370N5.3 |
| chr7_-_83824169 | 0.01 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr4_-_76598544 | 0.01 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr18_-_55288973 | 0.01 |
ENST00000423481.2
ENST00000587194.1 ENST00000591599.1 ENST00000588661.1 |
NARS
|
asparaginyl-tRNA synthetase |
| chr14_+_62462541 | 0.01 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr5_-_146258291 | 0.01 |
ENST00000394411.4
ENST00000453001.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chrX_-_73834449 | 0.01 |
ENST00000332687.6
ENST00000349225.2 |
RLIM
|
ring finger protein, LIM domain interacting |
| chr9_-_86323118 | 0.01 |
ENST00000376395.4
|
UBQLN1
|
ubiquilin 1 |
| chr1_-_168698433 | 0.01 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr4_-_10023095 | 0.01 |
ENST00000264784.3
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr3_+_73110810 | 0.01 |
ENST00000533473.1
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2 |
| chr11_-_119187826 | 0.01 |
ENST00000264036.4
|
MCAM
|
melanoma cell adhesion molecule |
| chr3_+_40498783 | 0.01 |
ENST00000338970.6
ENST00000396203.2 ENST00000416518.1 |
RPL14
|
ribosomal protein L14 |
| chr12_+_53835425 | 0.01 |
ENST00000549924.1
|
PRR13
|
proline rich 13 |
| chr6_-_30710447 | 0.01 |
ENST00000456573.2
|
FLOT1
|
flotillin 1 |
| chrX_+_47082408 | 0.01 |
ENST00000518022.1
ENST00000276052.6 |
CDK16
|
cyclin-dependent kinase 16 |
| chr3_-_194072019 | 0.01 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr7_-_83824449 | 0.01 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr13_-_46679144 | 0.01 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr17_-_76124711 | 0.01 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr4_+_86699834 | 0.01 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr13_-_46679185 | 0.01 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr3_-_157221128 | 0.01 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr14_+_39703084 | 0.01 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr17_+_34900737 | 0.01 |
ENST00000304718.4
ENST00000485685.2 |
GGNBP2
|
gametogenetin binding protein 2 |
| chr19_-_4540486 | 0.01 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr6_+_167704838 | 0.01 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr19_-_49496557 | 0.01 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr1_+_90308981 | 0.00 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr6_+_167704798 | 0.00 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr17_+_57287228 | 0.00 |
ENST00000578922.1
ENST00000300917.5 |
SMG8
|
SMG8 nonsense mediated mRNA decay factor |
| chr6_+_31895287 | 0.00 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr21_+_38792602 | 0.00 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr10_-_70092635 | 0.00 |
ENST00000309049.4
|
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr15_-_34628951 | 0.00 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr6_-_30710510 | 0.00 |
ENST00000376389.3
|
FLOT1
|
flotillin 1 |
| chr2_+_120303717 | 0.00 |
ENST00000594371.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
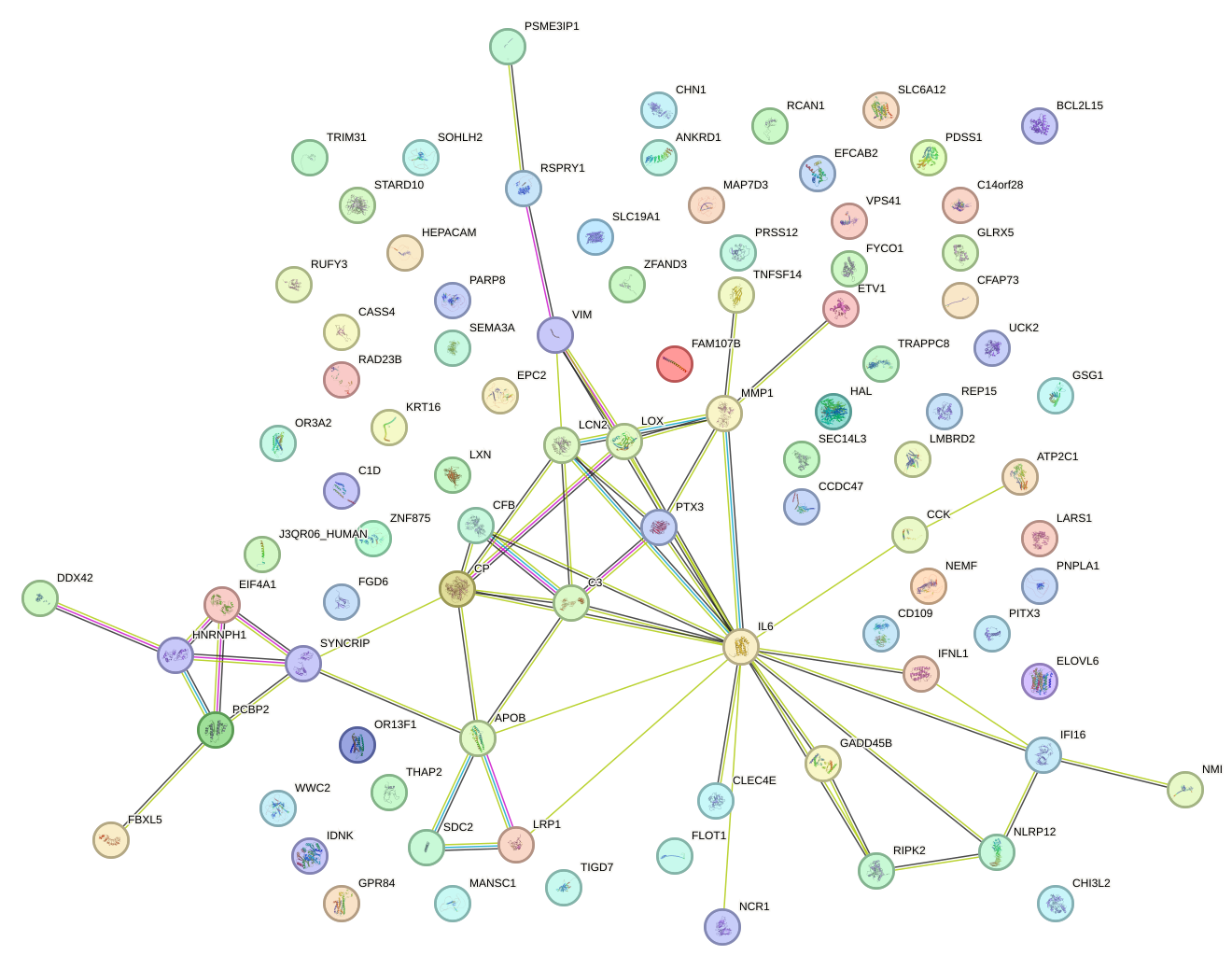
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.2 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |