Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
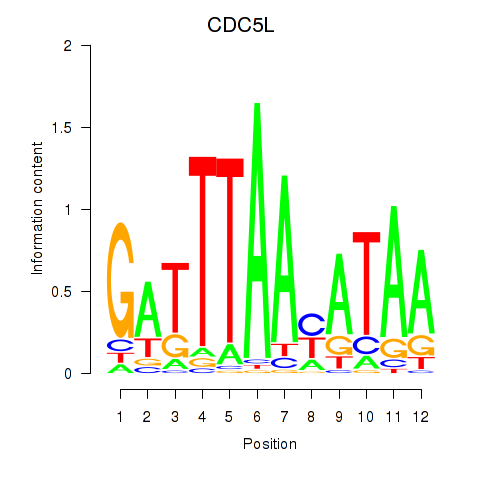
Results for CDC5L
Z-value: 0.73
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.7 | cell division cycle 5 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg19_v2_chr6_+_44355257_44355315 | 0.55 | 4.5e-01 | Click! |
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_11129229 | 0.44 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr2_+_200472779 | 0.37 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr18_-_37380230 | 0.31 |
ENST00000591629.1
|
LINC00669
|
long intergenic non-protein coding RNA 669 |
| chr1_-_154178803 | 0.30 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr3_+_57882061 | 0.29 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr14_+_104029362 | 0.29 |
ENST00000495778.1
|
APOPT1
|
apoptogenic 1, mitochondrial |
| chr21_+_35107346 | 0.28 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr6_+_76599809 | 0.27 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr8_+_52730143 | 0.27 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr2_-_203103185 | 0.24 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chr16_-_69418553 | 0.24 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr3_-_196987309 | 0.24 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr17_-_15469590 | 0.24 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr4_-_130692631 | 0.24 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr15_+_32933866 | 0.23 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr8_+_11666649 | 0.23 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr12_-_123756781 | 0.23 |
ENST00000544658.1
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr12_-_49463620 | 0.22 |
ENST00000550675.1
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chrX_-_45710920 | 0.21 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr18_+_20494078 | 0.21 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr10_-_69597915 | 0.21 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_+_124739964 | 0.20 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr14_+_39703112 | 0.20 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr17_+_18647326 | 0.20 |
ENST00000395667.1
ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chr22_-_40929812 | 0.20 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr10_-_69597828 | 0.20 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_+_9599540 | 0.19 |
ENST00000302692.6
|
SLC25A33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chrX_-_15402498 | 0.19 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr15_+_80364901 | 0.19 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr21_+_37692481 | 0.18 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr1_-_209741018 | 0.18 |
ENST00000424696.2
|
RP1-272L16.1
|
RP1-272L16.1 |
| chr4_+_74606223 | 0.18 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr12_-_25150409 | 0.17 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr4_+_80584903 | 0.17 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr15_+_35270552 | 0.16 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr15_+_67814008 | 0.16 |
ENST00000557807.1
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr9_-_16705069 | 0.16 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr20_+_52824367 | 0.16 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr21_+_17566643 | 0.15 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr11_-_92931098 | 0.15 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chrX_+_100646190 | 0.15 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr11_-_85430088 | 0.15 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr11_+_59705928 | 0.15 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr5_+_43033818 | 0.15 |
ENST00000607830.1
|
CTD-2035E11.4
|
CTD-2035E11.4 |
| chr15_+_49715449 | 0.15 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr12_+_16500599 | 0.14 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr10_-_69597810 | 0.14 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr8_+_76452097 | 0.14 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr1_-_65468148 | 0.14 |
ENST00000415842.1
|
RP11-182I10.3
|
RP11-182I10.3 |
| chr16_-_15149828 | 0.14 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr10_+_124739911 | 0.14 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr9_+_40028620 | 0.14 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr2_+_228678550 | 0.14 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr18_+_68002675 | 0.14 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr7_-_102985288 | 0.13 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr14_+_93260569 | 0.13 |
ENST00000163416.2
|
GOLGA5
|
golgin A5 |
| chr14_-_92247032 | 0.13 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr7_-_14026063 | 0.13 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr6_-_74233480 | 0.13 |
ENST00000455918.1
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr19_-_49843539 | 0.13 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr10_+_115511213 | 0.13 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr11_+_69924639 | 0.13 |
ENST00000538023.1
ENST00000398543.2 |
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr7_+_116654935 | 0.13 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr16_-_69419473 | 0.13 |
ENST00000566750.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr1_+_43613612 | 0.13 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr1_-_247094628 | 0.13 |
ENST00000366508.1
ENST00000326225.3 ENST00000391829.2 |
AHCTF1
|
AT hook containing transcription factor 1 |
| chrX_-_23926004 | 0.12 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr12_-_10542617 | 0.12 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chrX_-_15683147 | 0.12 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr7_-_102985035 | 0.12 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr8_-_102216925 | 0.12 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr12_-_122658746 | 0.12 |
ENST00000377035.1
|
IL31
|
interleukin 31 |
| chr11_-_46113756 | 0.12 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr3_-_119396193 | 0.12 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr6_+_161123270 | 0.12 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr2_-_203103281 | 0.12 |
ENST00000392244.3
ENST00000409181.1 ENST00000409712.1 ENST00000409498.2 ENST00000409368.1 ENST00000392245.1 ENST00000392246.2 |
SUMO1
|
small ubiquitin-like modifier 1 |
| chr10_+_27793197 | 0.12 |
ENST00000356940.6
ENST00000535776.1 |
RAB18
|
RAB18, member RAS oncogene family |
| chr8_-_101719159 | 0.12 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr11_+_115498761 | 0.12 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr19_+_30433110 | 0.12 |
ENST00000542441.2
ENST00000392271.1 |
URI1
|
URI1, prefoldin-like chaperone |
| chr6_+_13182751 | 0.12 |
ENST00000415087.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr3_+_108321623 | 0.12 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr17_-_7493390 | 0.12 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr1_-_86848760 | 0.12 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr12_+_20963647 | 0.12 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr19_+_37178482 | 0.12 |
ENST00000536254.2
|
ZNF567
|
zinc finger protein 567 |
| chr3_+_171561127 | 0.12 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr13_+_78315295 | 0.11 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_-_76912070 | 0.11 |
ENST00000395711.4
ENST00000356260.5 |
SDAD1
|
SDA1 domain containing 1 |
| chr12_-_49581152 | 0.11 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr16_+_66600294 | 0.11 |
ENST00000535705.1
ENST00000332695.7 ENST00000336328.6 ENST00000528324.1 ENST00000531885.1 ENST00000529506.1 ENST00000457188.2 ENST00000533666.1 ENST00000533953.1 ENST00000379500.2 ENST00000328020.6 |
CMTM1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr13_+_78315348 | 0.11 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr8_-_57906362 | 0.11 |
ENST00000262644.4
|
IMPAD1
|
inositol monophosphatase domain containing 1 |
| chr12_+_100897130 | 0.11 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr19_-_37157713 | 0.11 |
ENST00000592829.1
ENST00000591370.1 ENST00000588268.1 ENST00000360357.4 |
ZNF461
|
zinc finger protein 461 |
| chr10_+_35484053 | 0.11 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr1_+_38022572 | 0.11 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr2_-_180871780 | 0.11 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr2_+_196521845 | 0.11 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr6_-_75828774 | 0.11 |
ENST00000493109.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr3_-_149051444 | 0.11 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr2_+_132479948 | 0.11 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr16_-_69418649 | 0.11 |
ENST00000566257.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr12_+_19593515 | 0.11 |
ENST00000360995.4
|
AEBP2
|
AE binding protein 2 |
| chr3_+_150321068 | 0.10 |
ENST00000471696.1
ENST00000477889.1 ENST00000485923.1 |
SELT
|
Selenoprotein T |
| chr1_+_84609944 | 0.10 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_+_119200215 | 0.10 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr22_+_31795509 | 0.10 |
ENST00000331457.4
|
DRG1
|
developmentally regulated GTP binding protein 1 |
| chr16_-_71918033 | 0.10 |
ENST00000425432.1
ENST00000313565.6 ENST00000568666.1 ENST00000562797.1 ENST00000564134.1 |
ZNF821
|
zinc finger protein 821 |
| chr19_+_21579908 | 0.10 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr1_+_63249796 | 0.10 |
ENST00000443289.1
ENST00000317868.4 ENST00000371120.3 |
ATG4C
|
autophagy related 4C, cysteine peptidase |
| chr3_-_64009658 | 0.10 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr3_+_177545563 | 0.10 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr19_-_48753028 | 0.10 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr12_-_111358372 | 0.10 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr2_+_85132749 | 0.10 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr3_+_46616017 | 0.10 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr1_+_156338993 | 0.10 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr20_-_16554078 | 0.10 |
ENST00000354981.2
ENST00000355755.3 ENST00000378003.2 ENST00000408042.1 |
KIF16B
|
kinesin family member 16B |
| chr6_+_52535878 | 0.10 |
ENST00000211314.4
|
TMEM14A
|
transmembrane protein 14A |
| chr6_-_138833630 | 0.10 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr15_+_45028719 | 0.09 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr9_+_69252845 | 0.09 |
ENST00000354995.3
|
BX255923.1
|
HDCMB45P; Uncharacterized protein |
| chr2_+_65454863 | 0.09 |
ENST00000260641.5
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr8_+_92082424 | 0.09 |
ENST00000285420.4
ENST00000404789.3 |
OTUD6B
|
OTU domain containing 6B |
| chr2_+_196521903 | 0.09 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_+_229440129 | 0.09 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr1_+_156308403 | 0.09 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr1_+_78470530 | 0.09 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr1_+_24018269 | 0.09 |
ENST00000374550.3
|
RPL11
|
ribosomal protein L11 |
| chr14_+_39944025 | 0.09 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr6_+_123038689 | 0.09 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr10_+_27793257 | 0.09 |
ENST00000375802.3
|
RAB18
|
RAB18, member RAS oncogene family |
| chr13_+_111855414 | 0.09 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr4_+_130017268 | 0.09 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr15_-_94614049 | 0.08 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr12_-_53601000 | 0.08 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr1_+_41204506 | 0.08 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr14_-_53331239 | 0.08 |
ENST00000553663.1
|
FERMT2
|
fermitin family member 2 |
| chr1_+_62439037 | 0.08 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr1_+_110254850 | 0.08 |
ENST00000369812.5
ENST00000256593.3 ENST00000369813.1 |
GSTM5
|
glutathione S-transferase mu 5 |
| chr14_+_104029278 | 0.08 |
ENST00000472726.2
ENST00000409074.2 ENST00000440963.1 ENST00000556253.2 ENST00000247618.4 |
RP11-73M18.2
APOPT1
|
Kinesin light chain 1 apoptogenic 1, mitochondrial |
| chr5_+_95997769 | 0.08 |
ENST00000338252.3
ENST00000508830.1 |
CAST
|
calpastatin |
| chr15_+_81391740 | 0.08 |
ENST00000561216.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr16_-_27279900 | 0.08 |
ENST00000564342.1
ENST00000567710.1 ENST00000563273.1 ENST00000361439.4 |
NSMCE1
|
non-SMC element 1 homolog (S. cerevisiae) |
| chr5_+_179135246 | 0.08 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr8_+_99956759 | 0.08 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr7_+_116593433 | 0.08 |
ENST00000323984.3
ENST00000393449.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr9_-_21228221 | 0.08 |
ENST00000413767.2
|
IFNA17
|
interferon, alpha 17 |
| chr1_+_38022513 | 0.08 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr2_-_190927447 | 0.08 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr9_-_128412696 | 0.08 |
ENST00000420643.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr10_+_115511434 | 0.08 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr13_+_50589390 | 0.08 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr3_+_101443476 | 0.08 |
ENST00000327230.4
ENST00000494050.1 |
CEP97
|
centrosomal protein 97kDa |
| chr13_+_78315528 | 0.08 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_-_118959733 | 0.08 |
ENST00000459778.1
ENST00000359213.3 |
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr12_-_95044309 | 0.08 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr10_+_106113515 | 0.07 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr2_+_196521458 | 0.07 |
ENST00000409086.3
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr15_+_65823092 | 0.07 |
ENST00000566074.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr14_+_56127960 | 0.07 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr9_+_134165195 | 0.07 |
ENST00000372261.1
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr5_+_53686658 | 0.07 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr18_-_46784778 | 0.07 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr2_-_119605253 | 0.07 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr2_-_44550441 | 0.07 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr17_-_55911970 | 0.07 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr7_+_151038850 | 0.07 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr11_+_93479588 | 0.07 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr2_+_238877424 | 0.07 |
ENST00000434655.1
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr4_+_155484155 | 0.07 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr7_-_66460563 | 0.07 |
ENST00000246868.2
|
SBDS
|
Shwachman-Bodian-Diamond syndrome |
| chr5_-_43557791 | 0.07 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr5_-_179499108 | 0.07 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr5_+_118690466 | 0.07 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr3_-_150421728 | 0.07 |
ENST00000295910.6
ENST00000491361.1 |
FAM194A
|
family with sequence similarity 194, member A |
| chr15_-_55562479 | 0.07 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_+_111126707 | 0.07 |
ENST00000280325.4
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr5_-_157286104 | 0.07 |
ENST00000530742.1
ENST00000523908.1 ENST00000523094.1 ENST00000296951.5 ENST00000411809.2 |
CLINT1
|
clathrin interactor 1 |
| chr15_+_45028753 | 0.07 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr4_+_75023816 | 0.07 |
ENST00000395759.2
ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr4_+_48833015 | 0.07 |
ENST00000509122.1
ENST00000509664.1 ENST00000505922.2 ENST00000514981.1 |
OCIAD1
|
OCIA domain containing 1 |
| chrX_-_45629661 | 0.07 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr11_-_85430163 | 0.07 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr4_+_118349545 | 0.07 |
ENST00000422145.3
ENST00000437514.1 |
AC092661.1
|
AC092661.1 |
| chr7_+_142919130 | 0.07 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr12_+_16109519 | 0.07 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr1_-_115292591 | 0.07 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr14_-_38036271 | 0.07 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr1_+_63073437 | 0.07 |
ENST00000593719.1
|
AL138847.1
|
CDNA FLJ26506 fis, clone KDN07105; Uncharacterized protein |
| chr4_+_78829479 | 0.07 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr22_+_32871224 | 0.07 |
ENST00000452138.1
ENST00000382058.3 ENST00000397426.1 |
FBXO7
|
F-box protein 7 |
| chr22_+_19466980 | 0.07 |
ENST00000407835.1
ENST00000438587.1 |
CDC45
|
cell division cycle 45 |
| chr7_+_138943265 | 0.07 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr6_+_27356497 | 0.07 |
ENST00000244576.4
|
ZNF391
|
zinc finger protein 391 |
| chr10_-_16563870 | 0.06 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr2_-_197675000 | 0.06 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr3_-_156272924 | 0.06 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr11_-_31531121 | 0.06 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 0.5 | GO:1903824 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.3 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.3 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.2 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.3 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0045360 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.4 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0001079 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.1 | 0.3 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.4 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.5 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.0 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.0 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.0 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |