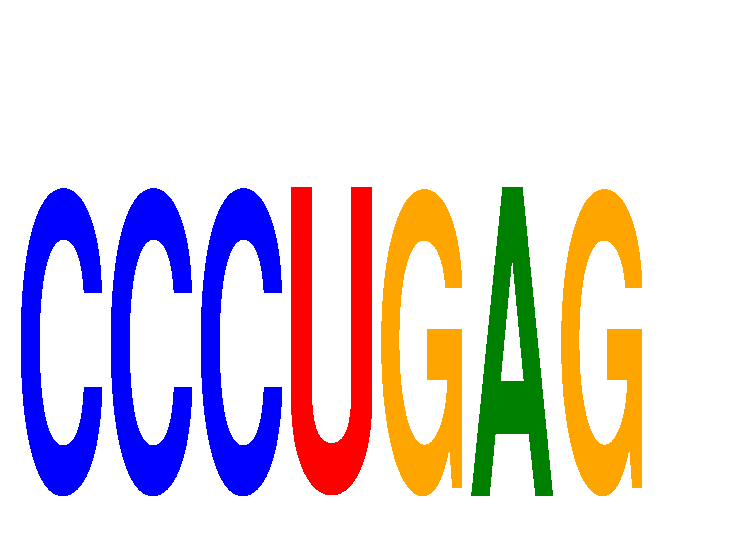Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CCCUGAG
Z-value: 2.02
miRNA associated with seed CCCUGAG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-125a-5p
|
MIMAT0000443 |
|
hsa-miR-125b-5p
|
MIMAT0000423 |
|
hsa-miR-4319
|
MIMAT0016870 |
Activity profile of CCCUGAG motif
Sorted Z-values of CCCUGAG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_1863567 | 1.53 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr11_-_65381643 | 1.19 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr19_+_7968728 | 1.03 |
ENST00000397981.3
ENST00000545011.1 ENST00000397983.3 ENST00000397979.3 |
MAP2K7
|
mitogen-activated protein kinase kinase 7 |
| chr17_-_61777459 | 0.98 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr11_+_64073699 | 0.96 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr17_-_40761375 | 0.87 |
ENST00000543197.1
ENST00000309428.5 |
FAM134C
|
family with sequence similarity 134, member C |
| chrX_-_48755030 | 0.80 |
ENST00000490755.2
ENST00000465150.2 ENST00000495490.2 |
TIMM17B
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr2_-_97535708 | 0.80 |
ENST00000305476.5
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr22_+_21771656 | 0.79 |
ENST00000407464.2
|
HIC2
|
hypermethylated in cancer 2 |
| chr20_-_33460621 | 0.78 |
ENST00000427420.1
ENST00000336431.5 |
GGT7
|
gamma-glutamyltransferase 7 |
| chr10_-_103454876 | 0.75 |
ENST00000331272.7
|
FBXW4
|
F-box and WD repeat domain containing 4 |
| chr20_-_32262165 | 0.72 |
ENST00000606690.1
ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3
|
N-terminal EF-hand calcium binding protein 3 |
| chr2_+_241375069 | 0.72 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr8_+_22857048 | 0.72 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr6_-_33547975 | 0.70 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr2_+_95963052 | 0.70 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chr19_+_42829702 | 0.68 |
ENST00000334370.4
|
MEGF8
|
multiple EGF-like-domains 8 |
| chr3_+_127391769 | 0.68 |
ENST00000393363.3
ENST00000232744.8 ENST00000453791.2 |
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr1_-_249120832 | 0.68 |
ENST00000366472.5
|
SH3BP5L
|
SH3-binding domain protein 5-like |
| chr15_-_75135453 | 0.67 |
ENST00000569437.1
ENST00000440863.2 |
ULK3
|
unc-51 like kinase 3 |
| chr20_+_2673383 | 0.65 |
ENST00000380648.4
ENST00000342725.5 |
EBF4
|
early B-cell factor 4 |
| chr19_-_4066890 | 0.65 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr17_-_78009647 | 0.62 |
ENST00000310924.2
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr10_-_73533255 | 0.62 |
ENST00000394957.3
|
C10orf54
|
chromosome 10 open reading frame 54 |
| chr2_+_220042933 | 0.62 |
ENST00000430297.2
|
FAM134A
|
family with sequence similarity 134, member A |
| chr22_-_39548627 | 0.61 |
ENST00000216133.5
|
CBX7
|
chromobox homolog 7 |
| chr2_+_74881355 | 0.61 |
ENST00000357877.2
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr19_+_45596218 | 0.61 |
ENST00000421905.1
ENST00000221462.4 |
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr19_+_10654561 | 0.60 |
ENST00000309469.4
|
ATG4D
|
autophagy related 4D, cysteine peptidase |
| chr12_-_50222187 | 0.60 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr2_+_28615669 | 0.60 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr17_-_8093471 | 0.59 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr12_-_56224546 | 0.58 |
ENST00000357606.3
ENST00000547445.1 |
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr6_-_31628512 | 0.57 |
ENST00000375911.1
|
C6orf47
|
chromosome 6 open reading frame 47 |
| chr17_+_73512594 | 0.57 |
ENST00000333213.6
|
TSEN54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr19_-_41196534 | 0.55 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr1_+_33207381 | 0.54 |
ENST00000401073.2
|
KIAA1522
|
KIAA1522 |
| chr19_+_13261216 | 0.54 |
ENST00000587885.1
ENST00000292433.3 |
IER2
|
immediate early response 2 |
| chr17_+_42836329 | 0.54 |
ENST00000200557.6
|
ADAM11
|
ADAM metallopeptidase domain 11 |
| chr8_+_37887772 | 0.54 |
ENST00000338825.4
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr19_-_11450249 | 0.53 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr5_+_10564432 | 0.51 |
ENST00000296657.5
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr16_+_4897632 | 0.51 |
ENST00000262376.6
|
UBN1
|
ubinuclein 1 |
| chrX_+_153639856 | 0.50 |
ENST00000426834.1
ENST00000369790.4 ENST00000454722.1 ENST00000350743.4 ENST00000299328.5 ENST00000351413.4 |
TAZ
|
tafazzin |
| chr11_+_45825896 | 0.50 |
ENST00000314134.3
|
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr11_-_46940074 | 0.49 |
ENST00000378623.1
ENST00000534404.1 |
LRP4
|
low density lipoprotein receptor-related protein 4 |
| chr19_+_926000 | 0.49 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr1_-_21671968 | 0.49 |
ENST00000415912.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr8_-_141645645 | 0.47 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr5_-_175964366 | 0.47 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr14_-_23504087 | 0.45 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr14_+_23775971 | 0.44 |
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2 |
| chr1_+_203274639 | 0.44 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr12_+_49932886 | 0.44 |
ENST00000257981.6
|
KCNH3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr9_-_127533519 | 0.44 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr19_+_13906250 | 0.44 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr8_-_81787006 | 0.43 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr1_-_36022979 | 0.43 |
ENST00000469892.1
ENST00000325722.3 |
KIAA0319L
|
KIAA0319-like |
| chr15_+_31619013 | 0.43 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr4_-_2264015 | 0.42 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr6_-_16761678 | 0.42 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr19_+_55851221 | 0.42 |
ENST00000255613.3
ENST00000539076.1 |
SUV420H2
AC020922.1
|
suppressor of variegation 4-20 homolog 2 (Drosophila) Uncharacterized protein |
| chr22_+_30279144 | 0.42 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr16_-_2264779 | 0.41 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr5_+_177019159 | 0.40 |
ENST00000332598.6
|
TMED9
|
transmembrane emp24 protein transport domain containing 9 |
| chr10_-_71906342 | 0.40 |
ENST00000287078.6
ENST00000335494.5 |
TYSND1
|
trypsin domain containing 1 |
| chr8_-_145550571 | 0.40 |
ENST00000332324.4
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr19_-_18632861 | 0.40 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr2_-_75788038 | 0.40 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr10_+_99258625 | 0.39 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr2_-_201936302 | 0.39 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr5_+_137673945 | 0.39 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr16_+_57126428 | 0.39 |
ENST00000290776.8
|
CPNE2
|
copine II |
| chr19_+_40697514 | 0.38 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr15_-_93199069 | 0.38 |
ENST00000327355.5
|
FAM174B
|
family with sequence similarity 174, member B |
| chr19_+_4969116 | 0.38 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr9_+_12775011 | 0.38 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr3_+_51575596 | 0.38 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr7_+_75831181 | 0.37 |
ENST00000388802.4
ENST00000326382.8 |
SRRM3
|
serine/arginine repetitive matrix 3 |
| chr12_-_54779511 | 0.36 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr17_+_72983674 | 0.35 |
ENST00000337231.5
|
CDR2L
|
cerebellar degeneration-related protein 2-like |
| chr19_-_15560730 | 0.35 |
ENST00000389282.4
ENST00000263381.7 |
WIZ
|
widely interspaced zinc finger motifs |
| chr3_+_126707437 | 0.35 |
ENST00000393409.2
ENST00000251772.4 |
PLXNA1
|
plexin A1 |
| chr11_-_62494821 | 0.35 |
ENST00000301785.5
|
HNRNPUL2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr10_+_102295616 | 0.35 |
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr8_-_28243934 | 0.35 |
ENST00000521185.1
ENST00000520290.1 ENST00000344423.5 |
ZNF395
|
zinc finger protein 395 |
| chr9_+_131102925 | 0.34 |
ENST00000372870.1
ENST00000300456.4 |
SLC27A4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr16_+_69221028 | 0.34 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr17_+_2240775 | 0.33 |
ENST00000268989.3
ENST00000426855.2 |
SGSM2
|
small G protein signaling modulator 2 |
| chr3_-_13461807 | 0.33 |
ENST00000254508.5
|
NUP210
|
nucleoporin 210kDa |
| chr17_+_38375574 | 0.33 |
ENST00000323571.4
ENST00000585043.1 ENST00000394103.3 ENST00000536600.1 |
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr2_+_47168313 | 0.33 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr3_-_133614597 | 0.33 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr11_+_94277017 | 0.32 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr19_+_50169081 | 0.32 |
ENST00000246784.3
|
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr3_+_47021168 | 0.32 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr9_+_139981379 | 0.32 |
ENST00000371589.4
|
MAN1B1
|
mannosidase, alpha, class 1B, member 1 |
| chr11_+_130318869 | 0.31 |
ENST00000299164.2
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr5_+_133861790 | 0.31 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr11_+_58939965 | 0.31 |
ENST00000227451.3
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr6_-_31830655 | 0.30 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr9_+_35538616 | 0.30 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr2_-_103353277 | 0.30 |
ENST00000258436.5
|
MFSD9
|
major facilitator superfamily domain containing 9 |
| chr6_-_32811771 | 0.29 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr11_+_63706444 | 0.29 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr10_+_134000404 | 0.29 |
ENST00000338492.4
ENST00000368629.1 |
DPYSL4
|
dihydropyrimidinase-like 4 |
| chr6_-_30658745 | 0.28 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chr11_+_76494253 | 0.28 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr22_+_41697520 | 0.28 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr1_+_29563011 | 0.28 |
ENST00000345512.3
ENST00000373779.3 ENST00000356870.3 ENST00000323874.8 ENST00000428026.2 ENST00000460170.2 |
PTPRU
|
protein tyrosine phosphatase, receptor type, U |
| chr1_+_109792641 | 0.28 |
ENST00000271332.3
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr1_+_20878932 | 0.28 |
ENST00000332947.4
|
FAM43B
|
family with sequence similarity 43, member B |
| chr11_+_61447845 | 0.28 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr8_-_8751068 | 0.28 |
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr1_+_151483855 | 0.27 |
ENST00000427934.2
ENST00000271636.7 |
CGN
|
cingulin |
| chr19_+_11485333 | 0.27 |
ENST00000312423.2
|
SWSAP1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr20_+_3827459 | 0.27 |
ENST00000416600.2
ENST00000428216.2 |
MAVS
|
mitochondrial antiviral signaling protein |
| chr2_+_175199674 | 0.27 |
ENST00000394967.2
|
SP9
|
Sp9 transcription factor |
| chr12_-_125348448 | 0.27 |
ENST00000339570.5
|
SCARB1
|
scavenger receptor class B, member 1 |
| chr17_-_21156578 | 0.27 |
ENST00000399011.2
ENST00000468196.1 |
C17orf103
|
chromosome 17 open reading frame 103 |
| chr11_-_64901978 | 0.26 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr14_+_70078303 | 0.26 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247 |
| chr16_-_28936493 | 0.26 |
ENST00000544477.1
ENST00000357573.6 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr10_-_15762124 | 0.26 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr12_+_56137064 | 0.26 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr11_-_78052923 | 0.26 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr9_-_130341268 | 0.25 |
ENST00000373314.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chr8_-_116681221 | 0.25 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr17_+_61699766 | 0.25 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr1_+_47901689 | 0.25 |
ENST00000334793.5
|
FOXD2
|
forkhead box D2 |
| chrX_-_128977781 | 0.24 |
ENST00000357166.6
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr3_-_39195037 | 0.24 |
ENST00000273153.5
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr20_+_55966444 | 0.24 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr1_+_32573636 | 0.24 |
ENST00000373625.3
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr2_+_232651124 | 0.24 |
ENST00000350033.3
ENST00000412591.1 ENST00000410017.1 ENST00000373608.3 |
COPS7B
|
COP9 signalosome subunit 7B |
| chr15_-_61521495 | 0.24 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr10_+_82213904 | 0.24 |
ENST00000429989.3
|
TSPAN14
|
tetraspanin 14 |
| chr16_+_1662326 | 0.24 |
ENST00000397412.3
|
CRAMP1L
|
Crm, cramped-like (Drosophila) |
| chr7_-_106301405 | 0.24 |
ENST00000523505.1
|
CCDC71L
|
coiled-coil domain containing 71-like |
| chr17_-_27507395 | 0.23 |
ENST00000354329.4
ENST00000527372.1 |
MYO18A
|
myosin XVIIIA |
| chr3_-_183273477 | 0.23 |
ENST00000341319.3
|
KLHL6
|
kelch-like family member 6 |
| chr8_-_66754172 | 0.23 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr9_+_77112244 | 0.23 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr12_+_104682496 | 0.23 |
ENST00000378070.4
|
TXNRD1
|
thioredoxin reductase 1 |
| chr17_-_41856305 | 0.23 |
ENST00000397937.2
ENST00000226004.3 |
DUSP3
|
dual specificity phosphatase 3 |
| chr9_+_34990219 | 0.23 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr17_-_79876010 | 0.23 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr6_+_43139037 | 0.22 |
ENST00000265354.4
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr2_+_85981008 | 0.22 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr18_+_18943554 | 0.22 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr18_-_77711625 | 0.22 |
ENST00000357575.4
ENST00000590381.1 ENST00000397778.2 |
PQLC1
|
PQ loop repeat containing 1 |
| chr1_-_23857698 | 0.22 |
ENST00000361729.2
|
E2F2
|
E2F transcription factor 2 |
| chr22_+_29279552 | 0.21 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr22_+_42229100 | 0.21 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr20_+_61436146 | 0.21 |
ENST00000290291.6
|
OGFR
|
opioid growth factor receptor |
| chr9_-_130829588 | 0.21 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr16_+_67063036 | 0.20 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr11_-_117103208 | 0.20 |
ENST00000320934.3
ENST00000530269.1 |
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr2_+_30454390 | 0.20 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr17_+_27717415 | 0.20 |
ENST00000583121.1
ENST00000261716.3 |
TAOK1
|
TAO kinase 1 |
| chr17_-_48207157 | 0.20 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr20_-_31071239 | 0.20 |
ENST00000359676.5
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr1_+_151584544 | 0.20 |
ENST00000458013.2
ENST00000368843.3 |
SNX27
|
sorting nexin family member 27 |
| chr14_+_56585048 | 0.20 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr17_-_76713100 | 0.20 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr11_+_65339820 | 0.20 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.1 |
FAM89B
|
family with sequence similarity 89, member B |
| chr4_-_71705590 | 0.20 |
ENST00000254799.6
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr1_+_205197304 | 0.20 |
ENST00000358024.3
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr1_+_160370344 | 0.20 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr9_-_14314066 | 0.19 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B |
| chr1_-_211307315 | 0.19 |
ENST00000271751.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chrX_+_78003204 | 0.19 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr17_+_27920486 | 0.19 |
ENST00000394859.3
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr3_+_42695176 | 0.19 |
ENST00000232974.6
ENST00000457842.3 |
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr5_+_139944024 | 0.19 |
ENST00000323146.3
|
SLC35A4
|
solute carrier family 35, member A4 |
| chr6_+_105404899 | 0.19 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chrX_-_103401649 | 0.19 |
ENST00000357421.4
|
SLC25A53
|
solute carrier family 25, member 53 |
| chr20_-_4982132 | 0.19 |
ENST00000338244.1
ENST00000424750.2 |
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr1_-_156252590 | 0.18 |
ENST00000361813.5
ENST00000368267.5 |
SMG5
|
SMG5 nonsense mediated mRNA decay factor |
| chr3_+_32859510 | 0.18 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr17_-_42908155 | 0.18 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr12_-_42538657 | 0.18 |
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr12_-_49351303 | 0.18 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr11_-_64612041 | 0.17 |
ENST00000342711.5
|
CDC42BPG
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chrX_+_48554986 | 0.17 |
ENST00000376687.3
ENST00000453214.2 |
SUV39H1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr10_+_101088836 | 0.17 |
ENST00000356713.4
|
CNNM1
|
cyclin M1 |
| chr1_-_6321035 | 0.17 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr9_-_130497565 | 0.17 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr16_+_27561449 | 0.17 |
ENST00000261588.4
|
KIAA0556
|
KIAA0556 |
| chr10_-_94003003 | 0.17 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr3_+_38495333 | 0.17 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr14_-_75179774 | 0.17 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr14_-_74253948 | 0.17 |
ENST00000394071.2
|
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr19_+_18794470 | 0.17 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr5_-_100238956 | 0.17 |
ENST00000231461.5
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr11_+_71640071 | 0.16 |
ENST00000533380.1
ENST00000393713.3 ENST00000545854.1 |
RNF121
|
ring finger protein 121 |
| chr12_+_122150646 | 0.16 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr11_-_65769594 | 0.16 |
ENST00000532707.1
ENST00000533544.1 ENST00000526451.1 ENST00000312234.2 ENST00000530462.1 ENST00000525767.1 ENST00000529964.1 ENST00000527249.1 |
EIF1AD
|
eukaryotic translation initiation factor 1A domain containing |
| chr1_+_16693578 | 0.16 |
ENST00000401088.4
ENST00000471507.1 ENST00000401089.3 ENST00000375590.3 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr6_+_35995488 | 0.16 |
ENST00000229795.3
|
MAPK14
|
mitogen-activated protein kinase 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CCCUGAG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 | 1.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.7 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 | 0.5 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.4 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.5 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.3 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.5 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.5 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 1.0 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.5 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.6 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.1 | 0.2 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.2 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.3 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.2 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.3 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.1 | 0.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.7 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.8 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.2 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.8 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.3 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.4 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.0 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.0 | 1.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.4 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.6 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.8 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.3 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.6 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.0 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.0 | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.3 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.2 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.3 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.3 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.7 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.3 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.5 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.4 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.5 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.2 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 0.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.4 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.2 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 1.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.2 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.3 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.2 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 1.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 4.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.8 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |