Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CCAGUGU
Z-value: 0.34
miRNA associated with seed CCAGUGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-199a-5p
|
MIMAT0000231 |
|
hsa-miR-199b-5p
|
MIMAT0000263 |
Activity profile of CCAGUGU motif
Sorted Z-values of CCAGUGU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_56092187 | 0.33 |
ENST00000325421.4
ENST00000592239.1 |
ZNF579
|
zinc finger protein 579 |
| chr11_-_65381643 | 0.23 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr8_+_144679984 | 0.22 |
ENST00000504548.2
ENST00000321385.3 |
TIGD5
|
tigger transposable element derived 5 |
| chr22_+_31031639 | 0.16 |
ENST00000343605.4
ENST00000300385.8 |
SLC35E4
|
solute carrier family 35, member E4 |
| chr17_+_74261277 | 0.14 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr17_-_61777459 | 0.14 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr22_-_37415475 | 0.13 |
ENST00000403892.3
ENST00000249042.3 ENST00000438203.1 |
TST
|
thiosulfate sulfurtransferase (rhodanese) |
| chr14_+_105267250 | 0.11 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr11_-_66115032 | 0.11 |
ENST00000311181.4
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr1_-_21671968 | 0.10 |
ENST00000415912.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr11_-_119599794 | 0.10 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr8_+_37553261 | 0.10 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr19_-_6459746 | 0.10 |
ENST00000301454.4
ENST00000334510.5 |
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr18_-_74207146 | 0.10 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr22_-_46373004 | 0.09 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chrX_+_48916497 | 0.09 |
ENST00000496529.2
ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120
|
coiled-coil domain containing 120 |
| chr11_+_61520075 | 0.09 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr19_+_45312347 | 0.09 |
ENST00000270233.6
ENST00000591520.1 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr16_+_11762270 | 0.09 |
ENST00000329565.5
|
SNN
|
stannin |
| chr19_+_12902289 | 0.09 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chr6_+_21593972 | 0.09 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr1_+_27561007 | 0.09 |
ENST00000319394.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr1_+_19970657 | 0.09 |
ENST00000375136.3
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr11_+_57227981 | 0.08 |
ENST00000335099.3
|
RTN4RL2
|
reticulon 4 receptor-like 2 |
| chr1_+_36348790 | 0.08 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr6_-_107436473 | 0.08 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr10_-_46030841 | 0.08 |
ENST00000453424.2
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr1_-_208417620 | 0.08 |
ENST00000367033.3
|
PLXNA2
|
plexin A2 |
| chr3_-_114790179 | 0.08 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr19_+_10654561 | 0.08 |
ENST00000309469.4
|
ATG4D
|
autophagy related 4D, cysteine peptidase |
| chr17_-_46682321 | 0.08 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr12_-_120687948 | 0.08 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr4_-_99579733 | 0.07 |
ENST00000305798.3
|
TSPAN5
|
tetraspanin 5 |
| chr20_-_36156125 | 0.07 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr2_-_201936302 | 0.07 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr12_-_56652111 | 0.07 |
ENST00000267116.7
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr19_+_39616410 | 0.07 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr11_-_46940074 | 0.07 |
ENST00000378623.1
ENST00000534404.1 |
LRP4
|
low density lipoprotein receptor-related protein 4 |
| chr19_-_11039261 | 0.07 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr22_-_46933067 | 0.07 |
ENST00000262738.3
ENST00000395964.1 |
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr19_-_16682987 | 0.07 |
ENST00000431408.1
ENST00000436553.2 ENST00000595753.1 |
SLC35E1
|
solute carrier family 35, member E1 |
| chr22_-_42017021 | 0.07 |
ENST00000263256.6
|
DESI1
|
desumoylating isopeptidase 1 |
| chr16_+_16043406 | 0.07 |
ENST00000399410.3
ENST00000399408.2 ENST00000346370.5 ENST00000351154.5 ENST00000345148.5 ENST00000349029.5 |
ABCC1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr20_-_62601218 | 0.07 |
ENST00000369888.1
|
ZNF512B
|
zinc finger protein 512B |
| chr19_+_7598890 | 0.07 |
ENST00000221249.6
ENST00000601668.1 ENST00000601001.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr11_-_62414070 | 0.06 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr11_+_63706444 | 0.06 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chrX_+_54466829 | 0.06 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr3_+_38495333 | 0.06 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr12_+_57482665 | 0.06 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr19_+_45349432 | 0.06 |
ENST00000252485.4
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr5_+_139493665 | 0.06 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr17_-_27916621 | 0.06 |
ENST00000225394.3
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr22_+_30279144 | 0.06 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr15_+_67813406 | 0.06 |
ENST00000342683.4
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr8_-_81787006 | 0.06 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr18_+_55102917 | 0.06 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr19_+_13906250 | 0.05 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr15_-_75135453 | 0.05 |
ENST00000569437.1
ENST00000440863.2 |
ULK3
|
unc-51 like kinase 3 |
| chr10_-_15762124 | 0.05 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr12_-_92539614 | 0.05 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr7_+_69064300 | 0.05 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr10_+_18549645 | 0.05 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr10_+_99496872 | 0.05 |
ENST00000337540.7
ENST00000357540.4 ENST00000370613.3 ENST00000370610.3 ENST00000393677.4 ENST00000453958.2 ENST00000359980.3 |
ZFYVE27
|
zinc finger, FYVE domain containing 27 |
| chr17_+_685513 | 0.05 |
ENST00000304478.4
|
RNMTL1
|
RNA methyltransferase like 1 |
| chr8_-_91997427 | 0.05 |
ENST00000517562.2
|
RP11-122A3.2
|
chromosome 8 open reading frame 88 |
| chr22_-_28197486 | 0.05 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr9_-_127952032 | 0.05 |
ENST00000456642.1
ENST00000373546.3 ENST00000373547.4 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr1_-_70820357 | 0.04 |
ENST00000370944.4
ENST00000262346.6 |
ANKRD13C
|
ankyrin repeat domain 13C |
| chr7_+_138916231 | 0.04 |
ENST00000473989.3
ENST00000288561.8 |
UBN2
|
ubinuclein 2 |
| chr8_+_37654424 | 0.04 |
ENST00000315215.7
|
GPR124
|
G protein-coupled receptor 124 |
| chr8_+_23386305 | 0.04 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr3_+_51575596 | 0.04 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr8_-_8751068 | 0.04 |
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr5_+_137774706 | 0.04 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr9_-_35732362 | 0.04 |
ENST00000314888.9
ENST00000540444.1 |
TLN1
|
talin 1 |
| chr12_-_54673871 | 0.04 |
ENST00000209875.4
|
CBX5
|
chromobox homolog 5 |
| chr16_-_30798492 | 0.04 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr19_+_49660997 | 0.04 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr9_+_77112244 | 0.04 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr22_-_18507279 | 0.04 |
ENST00000441493.2
ENST00000444520.1 ENST00000207726.7 ENST00000429452.1 |
MICAL3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr1_+_2985760 | 0.04 |
ENST00000378391.2
ENST00000514189.1 ENST00000270722.5 |
PRDM16
|
PR domain containing 16 |
| chr19_+_45754505 | 0.04 |
ENST00000262891.4
ENST00000300843.4 |
MARK4
|
MAP/microtubule affinity-regulating kinase 4 |
| chr3_-_129325660 | 0.03 |
ENST00000324093.4
ENST00000393239.1 |
PLXND1
|
plexin D1 |
| chr15_+_80987617 | 0.03 |
ENST00000258884.4
ENST00000558464.1 |
ABHD17C
|
abhydrolase domain containing 17C |
| chr11_+_77300669 | 0.03 |
ENST00000313578.3
|
AQP11
|
aquaporin 11 |
| chr17_+_4736627 | 0.03 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr20_+_34700333 | 0.03 |
ENST00000441639.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr5_-_59995921 | 0.03 |
ENST00000453022.2
ENST00000545085.1 ENST00000265036.5 |
DEPDC1B
|
DEP domain containing 1B |
| chr4_+_184826418 | 0.03 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr17_+_27717415 | 0.03 |
ENST00000583121.1
ENST00000261716.3 |
TAOK1
|
TAO kinase 1 |
| chr12_+_53574464 | 0.03 |
ENST00000416904.3
|
ZNF740
|
zinc finger protein 740 |
| chr10_-_62149433 | 0.03 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_+_53342311 | 0.03 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr9_-_36400213 | 0.03 |
ENST00000259605.6
ENST00000353739.4 |
RNF38
|
ring finger protein 38 |
| chr16_+_77224732 | 0.03 |
ENST00000569610.1
ENST00000248248.3 ENST00000567291.1 ENST00000320859.6 ENST00000563612.1 ENST00000563279.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr2_+_73429386 | 0.03 |
ENST00000398468.3
|
NOTO
|
notochord homeobox |
| chr16_-_23160591 | 0.03 |
ENST00000219689.7
|
USP31
|
ubiquitin specific peptidase 31 |
| chr9_+_35749203 | 0.03 |
ENST00000456972.2
ENST00000378078.4 |
RGP1
|
RGP1 retrograde golgi transport homolog (S. cerevisiae) |
| chr15_+_90544532 | 0.03 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr13_-_36050819 | 0.03 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr16_+_87636474 | 0.03 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr20_+_48429356 | 0.03 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_-_79479789 | 0.03 |
ENST00000571691.1
ENST00000571721.1 ENST00000573283.1 ENST00000575842.1 ENST00000575087.1 ENST00000570382.1 ENST00000331925.2 |
ACTG1
|
actin, gamma 1 |
| chr1_+_107683644 | 0.03 |
ENST00000370067.1
|
NTNG1
|
netrin G1 |
| chr17_-_26733179 | 0.02 |
ENST00000440501.1
ENST00000321666.5 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr14_+_71108460 | 0.02 |
ENST00000256367.2
|
TTC9
|
tetratricopeptide repeat domain 9 |
| chr3_+_50192537 | 0.02 |
ENST00000002829.3
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr7_+_100770328 | 0.02 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr3_-_183273477 | 0.02 |
ENST00000341319.3
|
KLHL6
|
kelch-like family member 6 |
| chr17_+_37026106 | 0.02 |
ENST00000318008.6
|
LASP1
|
LIM and SH3 protein 1 |
| chr22_+_41956767 | 0.02 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr12_+_104682496 | 0.02 |
ENST00000378070.4
|
TXNRD1
|
thioredoxin reductase 1 |
| chr1_+_160313062 | 0.02 |
ENST00000294785.5
ENST00000368063.1 ENST00000437169.1 |
NCSTN
|
nicastrin |
| chr20_-_25566153 | 0.02 |
ENST00000278886.6
ENST00000422516.1 |
NINL
|
ninein-like |
| chr9_+_34990219 | 0.02 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr10_+_102295616 | 0.02 |
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr1_+_244214577 | 0.02 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr3_-_15106747 | 0.02 |
ENST00000449354.2
ENST00000444840.2 ENST00000253686.2 |
MRPS25
|
mitochondrial ribosomal protein S25 |
| chr17_-_33448468 | 0.02 |
ENST00000591723.1
ENST00000593039.1 ENST00000587405.1 |
RAD51L3-RFFL
RAD51D
|
Uncharacterized protein RAD51 paralog D |
| chr8_-_66754172 | 0.02 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr2_-_19558373 | 0.02 |
ENST00000272223.2
|
OSR1
|
odd-skipped related transciption factor 1 |
| chr7_-_81399438 | 0.02 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr3_-_48471454 | 0.02 |
ENST00000296440.6
ENST00000448774.2 |
PLXNB1
|
plexin B1 |
| chr6_-_116601044 | 0.02 |
ENST00000368608.3
|
TSPYL1
|
TSPY-like 1 |
| chr19_-_56135928 | 0.02 |
ENST00000591479.1
ENST00000325351.4 |
ZNF784
|
zinc finger protein 784 |
| chr4_-_141075330 | 0.02 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr16_-_17564738 | 0.02 |
ENST00000261381.6
|
XYLT1
|
xylosyltransferase I |
| chr19_-_11308190 | 0.02 |
ENST00000586659.1
ENST00000592903.1 ENST00000589359.1 ENST00000588724.1 ENST00000432929.2 |
KANK2
|
KN motif and ankyrin repeat domains 2 |
| chr7_-_139763521 | 0.02 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr11_-_34379546 | 0.02 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr12_+_53774423 | 0.02 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr15_+_73976545 | 0.02 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr17_+_35849937 | 0.02 |
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14 |
| chr9_+_110045537 | 0.01 |
ENST00000358015.3
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr8_+_23430157 | 0.01 |
ENST00000399967.3
|
FP15737
|
FP15737 |
| chr22_-_44258360 | 0.01 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr13_-_30424821 | 0.01 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr19_+_38879061 | 0.01 |
ENST00000587013.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr10_+_89264625 | 0.01 |
ENST00000371996.4
ENST00000371994.4 |
MINPP1
|
multiple inositol-polyphosphate phosphatase 1 |
| chr7_+_140774032 | 0.01 |
ENST00000565468.1
|
TMEM178B
|
transmembrane protein 178B |
| chr14_+_20937538 | 0.01 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chr17_+_61086917 | 0.01 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr9_-_73029540 | 0.01 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr2_+_182321925 | 0.01 |
ENST00000339307.4
ENST00000397033.2 |
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
| chrX_+_49644470 | 0.01 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr12_+_52345448 | 0.01 |
ENST00000257963.4
ENST00000541224.1 ENST00000426655.2 ENST00000536420.1 ENST00000415850.2 |
ACVR1B
|
activin A receptor, type IB |
| chr4_-_53525406 | 0.01 |
ENST00000451218.2
ENST00000441222.3 |
USP46
|
ubiquitin specific peptidase 46 |
| chr14_-_39901618 | 0.01 |
ENST00000554932.1
ENST00000298097.7 |
FBXO33
|
F-box protein 33 |
| chr1_+_24742264 | 0.01 |
ENST00000374399.4
ENST00000003912.3 ENST00000358028.4 ENST00000339255.2 |
NIPAL3
|
NIPA-like domain containing 3 |
| chr14_+_50359773 | 0.01 |
ENST00000298316.5
|
ARF6
|
ADP-ribosylation factor 6 |
| chr1_+_38259540 | 0.01 |
ENST00000397631.3
|
MANEAL
|
mannosidase, endo-alpha-like |
| chrX_-_77150985 | 0.01 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr9_+_37422663 | 0.01 |
ENST00000318158.6
ENST00000607784.1 |
GRHPR
|
glyoxylate reductase/hydroxypyruvate reductase |
| chr11_+_119076745 | 0.01 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr19_-_10121144 | 0.01 |
ENST00000264828.3
|
COL5A3
|
collagen, type V, alpha 3 |
| chrX_-_54384425 | 0.01 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr9_-_19786926 | 0.01 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr4_+_145567173 | 0.01 |
ENST00000296575.3
|
HHIP
|
hedgehog interacting protein |
| chr5_+_178322893 | 0.01 |
ENST00000361362.2
ENST00000520660.1 ENST00000520805.1 |
ZFP2
|
ZFP2 zinc finger protein |
| chr1_+_28995231 | 0.01 |
ENST00000373816.1
|
GMEB1
|
glucocorticoid modulatory element binding protein 1 |
| chr15_-_75871589 | 0.01 |
ENST00000306726.2
|
PTPN9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr3_-_176914238 | 0.01 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr2_+_191273052 | 0.01 |
ENST00000417958.1
ENST00000432036.1 ENST00000392328.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr6_+_10556215 | 0.01 |
ENST00000316170.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr20_-_24973318 | 0.01 |
ENST00000447138.1
|
APMAP
|
adipocyte plasma membrane associated protein |
| chr4_-_40631859 | 0.01 |
ENST00000295971.7
ENST00000319592.4 |
RBM47
|
RNA binding motif protein 47 |
| chr15_-_42264702 | 0.01 |
ENST00000220325.4
|
EHD4
|
EH-domain containing 4 |
| chr12_-_9102549 | 0.01 |
ENST00000000412.3
|
M6PR
|
mannose-6-phosphate receptor (cation dependent) |
| chr11_+_121322832 | 0.01 |
ENST00000260197.7
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr17_+_46125707 | 0.01 |
ENST00000584137.1
ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chrX_-_99891796 | 0.01 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr9_+_105757590 | 0.01 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr11_+_62104897 | 0.01 |
ENST00000415229.2
ENST00000535727.1 ENST00000301776.5 |
ASRGL1
|
asparaginase like 1 |
| chr10_-_120514720 | 0.01 |
ENST00000369151.3
ENST00000340214.4 |
CACUL1
|
CDK2-associated, cullin domain 1 |
| chr12_+_119419294 | 0.01 |
ENST00000267260.4
|
SRRM4
|
serine/arginine repetitive matrix 4 |
| chr19_+_11071546 | 0.01 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr4_-_23891693 | 0.01 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr11_-_118661828 | 0.01 |
ENST00000264018.4
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr16_+_86600857 | 0.01 |
ENST00000320354.4
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr10_+_75504105 | 0.01 |
ENST00000535742.1
ENST00000546025.1 ENST00000345254.4 ENST00000540668.1 ENST00000339365.2 ENST00000411652.2 |
SEC24C
|
SEC24 family member C |
| chr16_-_4987065 | 0.01 |
ENST00000590782.2
ENST00000345988.2 |
PPL
|
periplakin |
| chr1_+_15943995 | 0.01 |
ENST00000480945.1
|
DDI2
|
DNA-damage inducible 1 homolog 2 (S. cerevisiae) |
| chr2_-_176948641 | 0.01 |
ENST00000308618.4
|
EVX2
|
even-skipped homeobox 2 |
| chr20_+_19867150 | 0.01 |
ENST00000255006.6
|
RIN2
|
Ras and Rab interactor 2 |
| chr5_-_176778523 | 0.01 |
ENST00000513877.1
ENST00000515209.1 ENST00000514458.1 ENST00000502560.1 |
LMAN2
|
lectin, mannose-binding 2 |
| chr12_+_109535373 | 0.01 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chr4_+_6911171 | 0.01 |
ENST00000448507.1
|
TBC1D14
|
TBC1 domain family, member 14 |
| chr10_-_98346801 | 0.01 |
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr6_+_30951487 | 0.01 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr1_+_36396677 | 0.01 |
ENST00000373191.4
ENST00000397828.2 |
AGO3
|
argonaute RISC catalytic component 3 |
| chr15_-_63674218 | 0.00 |
ENST00000178638.3
|
CA12
|
carbonic anhydrase XII |
| chr7_-_27196267 | 0.00 |
ENST00000242159.3
|
HOXA7
|
homeobox A7 |
| chr14_+_103388976 | 0.00 |
ENST00000299155.5
|
AMN
|
amnion associated transmembrane protein |
| chr8_-_19459993 | 0.00 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr9_-_104145795 | 0.00 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr6_-_8435706 | 0.00 |
ENST00000379660.4
|
SLC35B3
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
| chr2_-_11606275 | 0.00 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr2_-_213403565 | 0.00 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr16_-_71843047 | 0.00 |
ENST00000299980.4
ENST00000393512.3 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
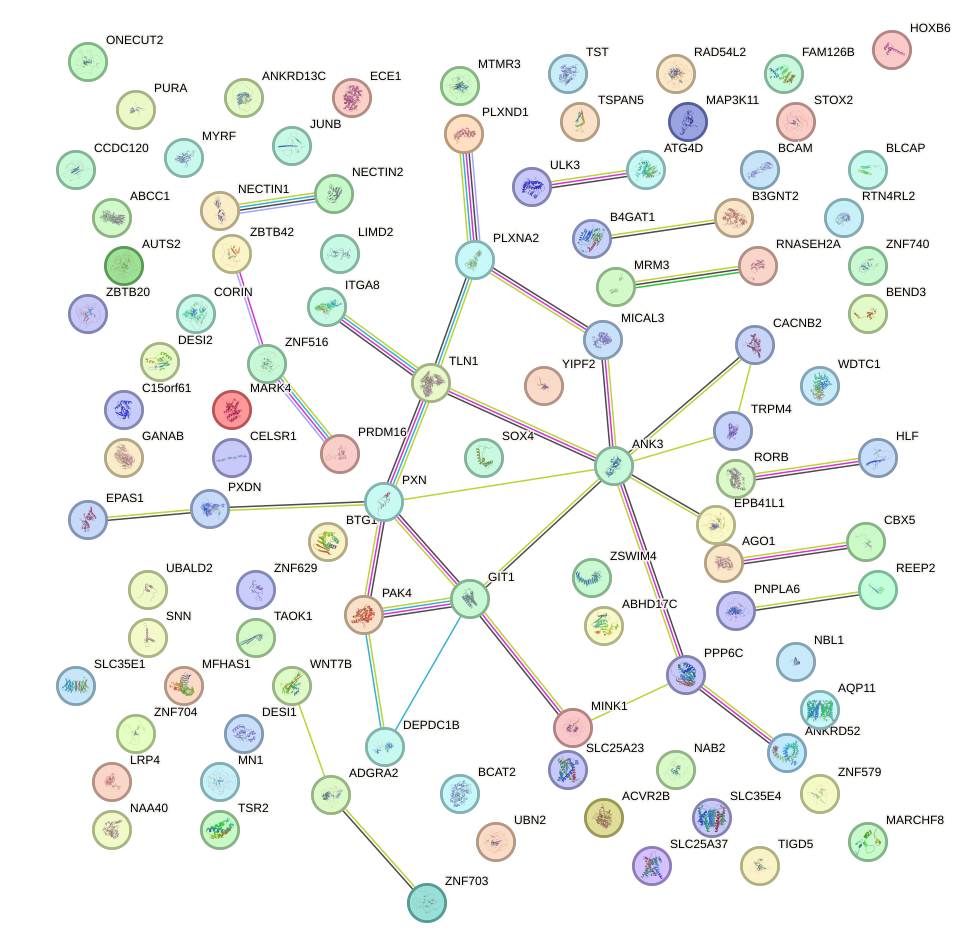
Network of associatons between targets according to the STRING database.
First level regulatory network of CCAGUGU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |