Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
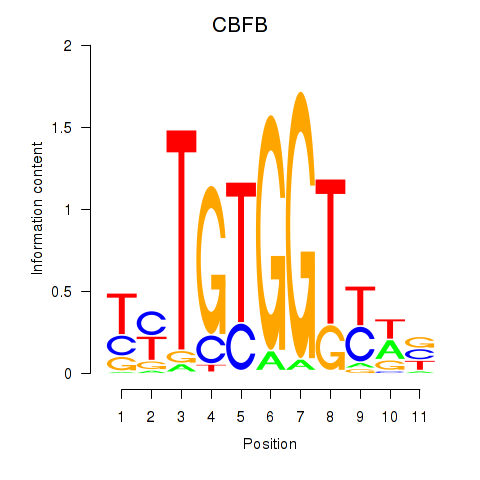
Results for CBFB
Z-value: 0.17
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | core-binding factor subunit beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg19_v2_chr16_+_67062996_67063019 | -0.80 | 2.0e-01 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_35107346 | 0.09 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr20_+_54987168 | 0.08 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr20_+_54987305 | 0.08 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr22_+_42665742 | 0.07 |
ENST00000332965.3
ENST00000415205.1 ENST00000446578.1 |
Z83851.3
|
Z83851.3 |
| chr1_+_17634689 | 0.06 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr1_-_52499443 | 0.05 |
ENST00000371614.1
|
KTI12
|
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr14_-_85996332 | 0.04 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr9_-_35079911 | 0.04 |
ENST00000448890.1
|
FANCG
|
Fanconi anemia, complementation group G |
| chr19_-_14049184 | 0.04 |
ENST00000339560.5
|
PODNL1
|
podocan-like 1 |
| chr17_+_32582293 | 0.04 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr21_-_36421626 | 0.03 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr12_-_121972556 | 0.03 |
ENST00000545022.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr21_-_36421535 | 0.03 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr2_-_127977654 | 0.03 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr3_+_98482175 | 0.03 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr3_-_99569821 | 0.03 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr15_+_49715293 | 0.02 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr19_-_14048804 | 0.02 |
ENST00000254320.3
ENST00000586075.1 |
PODNL1
|
podocan-like 1 |
| chr20_+_44637526 | 0.02 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr12_-_4754356 | 0.02 |
ENST00000540967.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr12_-_54778444 | 0.02 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr11_-_66103867 | 0.02 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr8_-_134072593 | 0.02 |
ENST00000427060.2
|
SLA
|
Src-like-adaptor |
| chr12_-_53594227 | 0.02 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chrX_+_115567767 | 0.02 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr6_+_30908747 | 0.02 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr3_-_11610255 | 0.02 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr11_-_66103932 | 0.02 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr7_+_7196565 | 0.01 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr14_-_61116168 | 0.01 |
ENST00000247182.6
|
SIX1
|
SIX homeobox 1 |
| chr12_-_4754339 | 0.01 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr12_-_54778244 | 0.01 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr19_+_10541462 | 0.01 |
ENST00000293683.5
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr11_-_66104237 | 0.01 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr13_-_103019744 | 0.01 |
ENST00000437115.2
|
FGF14-IT1
|
FGF14 intronic transcript 1 (non-protein coding) |
| chr21_-_36421401 | 0.01 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr17_+_1633755 | 0.01 |
ENST00000545662.1
|
WDR81
|
WD repeat domain 81 |
| chrX_-_19688475 | 0.01 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr1_-_179112189 | 0.01 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr1_-_184723942 | 0.01 |
ENST00000318130.8
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr15_+_67390920 | 0.01 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr1_+_66796401 | 0.01 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr1_+_39876151 | 0.01 |
ENST00000530275.1
|
KIAA0754
|
KIAA0754 |
| chr9_-_35080013 | 0.01 |
ENST00000378643.3
|
FANCG
|
Fanconi anemia, complementation group G |
| chr11_+_18343800 | 0.00 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr14_-_59951112 | 0.00 |
ENST00000247194.4
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr3_-_189840223 | 0.00 |
ENST00000427335.2
|
LEPREL1
|
leprecan-like 1 |
| chr8_-_28747424 | 0.00 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
| chr1_+_89246647 | 0.00 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr12_-_54778471 | 0.00 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr1_+_209859510 | 0.00 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr2_+_234826016 | 0.00 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr15_-_64665911 | 0.00 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr14_+_85996507 | 0.00 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr11_-_18343669 | 0.00 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr11_-_84028180 | 0.00 |
ENST00000280241.8
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chrX_-_153718988 | 0.00 |
ENST00000263512.4
ENST00000393587.4 ENST00000453912.1 |
SLC10A3
|
solute carrier family 10, member 3 |
| chr14_-_35183886 | 0.00 |
ENST00000298159.6
|
CFL2
|
cofilin 2 (muscle) |
| chr15_-_33447055 | 0.00 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chrX_-_49041242 | 0.00 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr18_-_53089538 | 0.00 |
ENST00000566777.1
|
TCF4
|
transcription factor 4 |
| chr11_-_18343725 | 0.00 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr6_-_152623231 | 0.00 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr20_+_62492566 | 0.00 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr4_+_74347400 | 0.00 |
ENST00000226355.3
|
AFM
|
afamin |
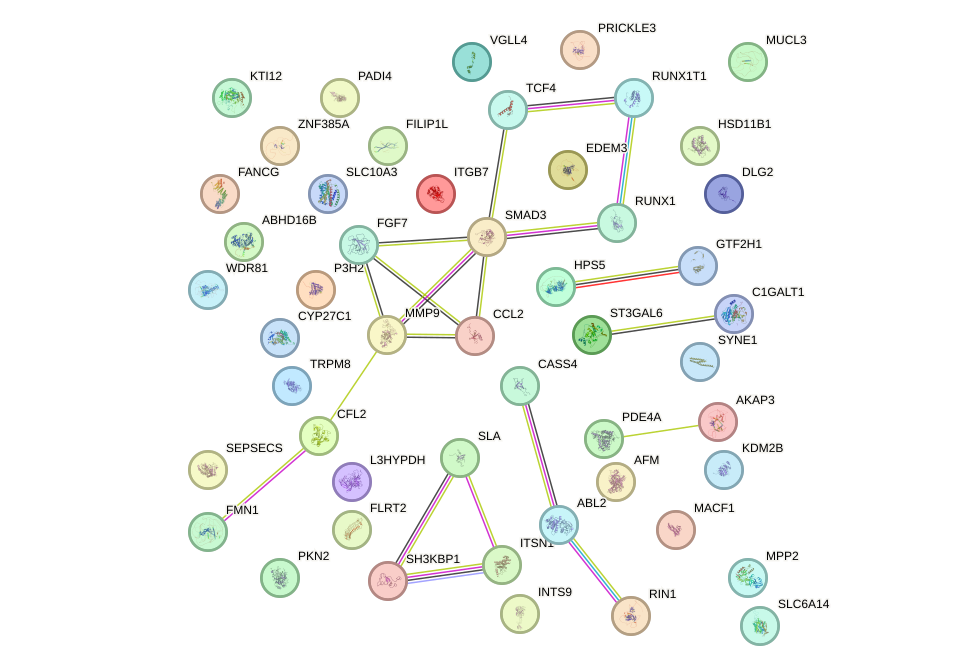
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |