Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
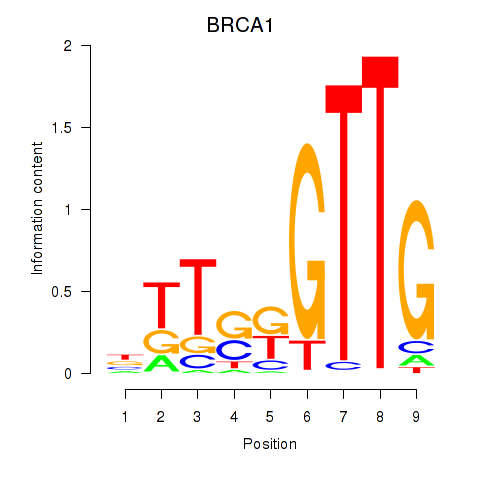
Results for BRCA1
Z-value: 0.26
Transcription factors associated with BRCA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BRCA1
|
ENSG00000012048.15 | BRCA1 DNA repair associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BRCA1 | hg19_v2_chr17_-_41277317_41277348 | 0.95 | 4.8e-02 | Click! |
Activity profile of BRCA1 motif
Sorted Z-values of BRCA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_67141640 | 0.17 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr8_+_37553261 | 0.14 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chrX_-_70288234 | 0.13 |
ENST00000276105.3
ENST00000374274.3 |
SNX12
|
sorting nexin 12 |
| chr17_+_21730180 | 0.12 |
ENST00000584398.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr19_-_40324767 | 0.10 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr17_-_27230035 | 0.09 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr14_+_32030582 | 0.09 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr14_-_69261310 | 0.08 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr12_-_49449107 | 0.06 |
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr10_+_104404644 | 0.06 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr5_+_139493665 | 0.06 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr17_-_56296580 | 0.06 |
ENST00000313863.6
ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1
|
Meckel syndrome, type 1 |
| chr2_+_196522032 | 0.06 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chrX_-_77150911 | 0.06 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr17_+_36584662 | 0.06 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr3_+_184056614 | 0.05 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr2_+_108994466 | 0.05 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr1_+_70687137 | 0.05 |
ENST00000436161.2
|
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr7_+_99717230 | 0.05 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr5_+_140186647 | 0.05 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr12_-_49418407 | 0.05 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr1_-_160232197 | 0.04 |
ENST00000419626.1
ENST00000610139.1 ENST00000475733.1 ENST00000407642.2 ENST00000368073.3 ENST00000326837.2 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr8_-_116681123 | 0.04 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr16_-_790982 | 0.04 |
ENST00000301694.5
ENST00000251588.2 |
NARFL
|
nuclear prelamin A recognition factor-like |
| chr7_-_99716952 | 0.04 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr3_-_127455200 | 0.04 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr10_-_75385711 | 0.04 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr6_+_42981922 | 0.04 |
ENST00000326974.4
ENST00000244670.8 |
KLHDC3
|
kelch domain containing 3 |
| chrX_+_150565038 | 0.04 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr8_+_22462532 | 0.03 |
ENST00000389279.3
|
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr10_+_102295616 | 0.03 |
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr6_+_12717892 | 0.03 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr9_-_86593238 | 0.03 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr10_-_102289611 | 0.03 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr19_+_45394477 | 0.03 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr1_-_160232291 | 0.03 |
ENST00000368074.1
ENST00000447377.1 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr6_-_42981651 | 0.03 |
ENST00000244711.3
|
MEA1
|
male-enhanced antigen 1 |
| chr8_-_116681221 | 0.03 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr5_-_124082279 | 0.03 |
ENST00000513986.1
|
ZNF608
|
zinc finger protein 608 |
| chr15_+_67430339 | 0.03 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr6_-_25832254 | 0.03 |
ENST00000476801.1
ENST00000244527.4 ENST00000427328.1 |
SLC17A1
|
solute carrier family 17 (organic anion transporter), member 1 |
| chr8_+_97773202 | 0.03 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr15_-_88799948 | 0.03 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr6_-_137314371 | 0.03 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr16_-_790887 | 0.02 |
ENST00000540986.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chr19_-_37663572 | 0.02 |
ENST00000588354.1
ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A
|
zinc finger protein 585A |
| chr17_+_74372662 | 0.02 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr19_-_48389651 | 0.02 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr14_-_75179774 | 0.02 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr1_-_173020056 | 0.02 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr2_-_179914760 | 0.02 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr10_-_61900762 | 0.02 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr10_+_122610853 | 0.02 |
ENST00000604585.1
|
WDR11
|
WD repeat domain 11 |
| chr6_+_33172407 | 0.02 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr11_+_47291193 | 0.02 |
ENST00000428807.1
ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD
|
MAP-kinase activating death domain |
| chr19_-_37663332 | 0.02 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr13_+_27825446 | 0.02 |
ENST00000311549.6
|
RPL21
|
ribosomal protein L21 |
| chrX_+_10124977 | 0.02 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr8_+_22022800 | 0.02 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr3_+_185303962 | 0.01 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr7_-_99716940 | 0.01 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr8_+_22022653 | 0.01 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr1_-_170043709 | 0.01 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr5_+_139027877 | 0.01 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr6_-_170862322 | 0.01 |
ENST00000262193.6
|
PSMB1
|
proteasome (prosome, macropain) subunit, beta type, 1 |
| chr10_+_122610687 | 0.01 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chrX_+_49644470 | 0.01 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr7_+_44646177 | 0.01 |
ENST00000443864.2
ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr7_+_44646162 | 0.01 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr19_+_46000479 | 0.01 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr17_+_36908984 | 0.01 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr1_+_10271674 | 0.01 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr12_+_122880045 | 0.01 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr1_-_115301235 | 0.00 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr3_-_176914191 | 0.00 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr2_-_158182410 | 0.00 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr2_-_158182105 | 0.00 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr1_+_70687079 | 0.00 |
ENST00000395136.2
|
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr8_+_97773457 | 0.00 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr2_-_158182322 | 0.00 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr19_+_55888186 | 0.00 |
ENST00000291934.3
|
TMEM190
|
transmembrane protein 190 |
| chr2_-_152118352 | 0.00 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr10_-_95209 | 0.00 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr2_+_203879568 | 0.00 |
ENST00000449802.1
|
NBEAL1
|
neurobeachin-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BRCA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |