Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
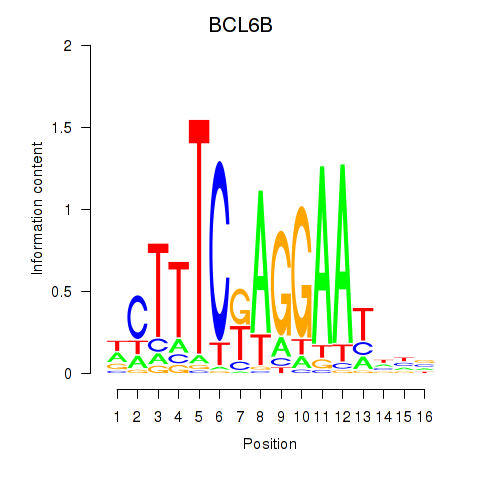
Results for BCL6B
Z-value: 0.47
Transcription factors associated with BCL6B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BCL6B
|
ENSG00000161940.6 | BCL6B transcription repressor |
Activity profile of BCL6B motif
Sorted Z-values of BCL6B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39786962 | 0.24 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr10_+_81065975 | 0.24 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr5_+_72251857 | 0.18 |
ENST00000507345.2
ENST00000512348.1 ENST00000287761.6 |
FCHO2
|
FCH domain only 2 |
| chr12_-_42877726 | 0.16 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr8_+_67039278 | 0.15 |
ENST00000276573.7
ENST00000350034.4 |
TRIM55
|
tripartite motif containing 55 |
| chr1_-_193029192 | 0.14 |
ENST00000417752.1
ENST00000367452.4 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr13_-_36705425 | 0.14 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr8_+_67039131 | 0.14 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr12_-_122985067 | 0.13 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr14_+_21566980 | 0.13 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr16_+_58283814 | 0.13 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr12_-_42877764 | 0.13 |
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr18_-_66382289 | 0.12 |
ENST00000443099.2
ENST00000562706.1 ENST00000544714.2 |
TMX3
|
thioredoxin-related transmembrane protein 3 |
| chr1_-_150669604 | 0.12 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr14_+_39944025 | 0.11 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr12_+_104324112 | 0.11 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr2_+_169659121 | 0.11 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr2_+_27805880 | 0.11 |
ENST00000379717.1
ENST00000355467.4 ENST00000556601.1 ENST00000416005.2 |
ZNF512
|
zinc finger protein 512 |
| chr5_+_72251793 | 0.11 |
ENST00000430046.2
ENST00000341845.6 |
FCHO2
|
FCH domain only 2 |
| chr12_-_11002063 | 0.11 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr14_-_30396948 | 0.10 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr3_-_23958402 | 0.10 |
ENST00000415901.2
ENST00000416026.2 ENST00000412028.1 ENST00000388759.3 ENST00000437230.1 |
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr11_+_107643129 | 0.10 |
ENST00000447610.1
|
AP001024.2
|
Uncharacterized protein |
| chr5_-_145214848 | 0.10 |
ENST00000505416.1
ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr13_-_73356009 | 0.10 |
ENST00000377780.4
ENST00000377767.4 |
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr2_+_27805971 | 0.10 |
ENST00000413371.2
|
ZNF512
|
zinc finger protein 512 |
| chr21_+_17102311 | 0.10 |
ENST00000285679.6
ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25
|
ubiquitin specific peptidase 25 |
| chr20_+_11873141 | 0.10 |
ENST00000422390.1
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr17_+_79761997 | 0.10 |
ENST00000400723.3
ENST00000570996.1 |
GCGR
|
glucagon receptor |
| chrX_+_95939638 | 0.09 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr9_+_70971815 | 0.09 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr4_-_99578776 | 0.09 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr20_-_18447667 | 0.09 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr2_+_169658928 | 0.09 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr15_-_38519066 | 0.08 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr13_+_73356197 | 0.08 |
ENST00000326291.6
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr14_+_31028348 | 0.08 |
ENST00000550944.1
ENST00000438909.2 ENST00000553504.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr9_-_113018835 | 0.08 |
ENST00000374517.5
|
TXN
|
thioredoxin |
| chr2_-_188419200 | 0.08 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_+_70121075 | 0.08 |
ENST00000409116.1
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chr13_-_73356234 | 0.08 |
ENST00000545453.1
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr2_-_55237484 | 0.08 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr14_+_31028329 | 0.08 |
ENST00000206595.6
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr2_-_157186630 | 0.08 |
ENST00000406048.2
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr13_-_33112823 | 0.08 |
ENST00000504114.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr11_+_58874701 | 0.07 |
ENST00000529618.1
ENST00000534403.1 ENST00000343597.3 |
FAM111B
|
family with sequence similarity 111, member B |
| chr15_+_74610894 | 0.07 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr12_+_16500037 | 0.07 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr2_-_188419078 | 0.07 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr6_-_45544507 | 0.07 |
ENST00000563807.1
|
RP1-166H4.2
|
RP1-166H4.2 |
| chr21_+_44394742 | 0.07 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr4_-_184241927 | 0.07 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr3_-_23958506 | 0.07 |
ENST00000425478.2
|
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr8_-_102216925 | 0.07 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr12_-_122985494 | 0.07 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr14_-_80678512 | 0.07 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr15_+_45003675 | 0.06 |
ENST00000558401.1
ENST00000559916.1 ENST00000544417.1 |
B2M
|
beta-2-microglobulin |
| chr11_+_73358690 | 0.06 |
ENST00000545798.1
ENST00000539157.1 ENST00000546251.1 ENST00000535582.1 ENST00000538227.1 ENST00000543524.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr1_-_108735440 | 0.06 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chrX_+_95939711 | 0.06 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr11_-_85430356 | 0.06 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr4_+_89378261 | 0.06 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr5_-_145214893 | 0.06 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr3_-_50649192 | 0.06 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr17_+_66539369 | 0.06 |
ENST00000600820.1
|
AC079210.1
|
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr16_-_66952779 | 0.06 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr4_+_95972822 | 0.06 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr13_-_28194541 | 0.06 |
ENST00000316334.3
|
LNX2
|
ligand of numb-protein X 2 |
| chr11_+_74303612 | 0.06 |
ENST00000527458.1
ENST00000532497.1 ENST00000530511.1 |
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr13_-_33112899 | 0.06 |
ENST00000267068.3
ENST00000357505.6 ENST00000399396.3 |
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr16_-_66952742 | 0.05 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr11_-_128775592 | 0.05 |
ENST00000310799.3
|
C11orf45
|
chromosome 11 open reading frame 45 |
| chr7_+_134464414 | 0.05 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr5_+_75904918 | 0.05 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr12_+_70132632 | 0.05 |
ENST00000378815.6
ENST00000483530.2 ENST00000325555.9 |
RAB3IP
|
RAB3A interacting protein |
| chr2_+_87754887 | 0.05 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr7_+_134464376 | 0.04 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr9_+_88556036 | 0.04 |
ENST00000361671.5
ENST00000416045.1 |
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr6_+_7541808 | 0.04 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr16_+_16481598 | 0.04 |
ENST00000327792.5
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr17_+_76183398 | 0.04 |
ENST00000409257.5
|
AFMID
|
arylformamidase |
| chr1_+_235530675 | 0.04 |
ENST00000366601.3
ENST00000406207.1 ENST00000543662.1 |
TBCE
|
tubulin folding cofactor E |
| chr13_-_41345277 | 0.04 |
ENST00000323563.6
|
MRPS31
|
mitochondrial ribosomal protein S31 |
| chr1_-_150669500 | 0.04 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr15_-_49913126 | 0.04 |
ENST00000561064.1
ENST00000299338.6 |
FAM227B
|
family with sequence similarity 227, member B |
| chr1_-_212004090 | 0.04 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr17_-_74489215 | 0.04 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr6_+_7541845 | 0.04 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chrX_-_139866723 | 0.04 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr10_-_73976884 | 0.04 |
ENST00000317126.4
ENST00000545550.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr11_-_128775930 | 0.04 |
ENST00000524878.1
|
C11orf45
|
chromosome 11 open reading frame 45 |
| chr7_+_74072011 | 0.04 |
ENST00000324896.4
ENST00000353920.4 ENST00000346152.4 ENST00000416070.1 |
GTF2I
|
general transcription factor IIi |
| chr17_-_54893250 | 0.04 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr6_+_35995552 | 0.04 |
ENST00000468133.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr6_+_150690133 | 0.03 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr11_+_73358594 | 0.03 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr19_-_44860820 | 0.03 |
ENST00000354340.4
ENST00000337401.4 ENST00000587909.1 |
ZNF112
|
zinc finger protein 112 |
| chr20_+_56136136 | 0.03 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr12_+_70132458 | 0.03 |
ENST00000247833.7
|
RAB3IP
|
RAB3A interacting protein |
| chr3_+_23958470 | 0.03 |
ENST00000434031.2
ENST00000413699.1 ENST00000456530.2 |
RPL15
|
ribosomal protein L15 |
| chr3_-_64673289 | 0.03 |
ENST00000295903.4
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr2_+_87754989 | 0.03 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr3_-_69101461 | 0.03 |
ENST00000543976.1
|
TMF1
|
TATA element modulatory factor 1 |
| chr3_+_177534653 | 0.03 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr1_+_84609944 | 0.03 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr19_+_45147313 | 0.03 |
ENST00000406449.4
|
PVR
|
poliovirus receptor |
| chr10_-_73976025 | 0.03 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chrX_-_57147748 | 0.03 |
ENST00000374910.3
|
SPIN2B
|
spindlin family, member 2B |
| chr15_+_26147507 | 0.03 |
ENST00000383019.2
ENST00000557171.1 |
RP11-1084I9.1
|
RP11-1084I9.1 |
| chr9_+_97766469 | 0.03 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr18_-_46895066 | 0.03 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr3_+_185046676 | 0.03 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chrX_-_134936550 | 0.03 |
ENST00000494421.1
|
CT45A4
|
cancer/testis antigen family 45, member A4 |
| chr15_+_69854027 | 0.03 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr21_+_17909594 | 0.03 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr13_-_41768654 | 0.03 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr7_+_872107 | 0.03 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr3_-_123603137 | 0.03 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr10_-_62493223 | 0.03 |
ENST00000373827.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr3_+_46618727 | 0.02 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chrX_+_134847370 | 0.02 |
ENST00000482795.1
|
CT45A1
|
cancer/testis antigen family 45, member A1 |
| chr20_+_58251716 | 0.02 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr9_-_117880477 | 0.02 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr12_+_79371565 | 0.02 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr11_+_64059464 | 0.02 |
ENST00000394525.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr4_+_86396321 | 0.02 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr5_-_58295712 | 0.02 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_48276710 | 0.02 |
ENST00000550314.1
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr19_-_4717835 | 0.02 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr11_+_58912240 | 0.02 |
ENST00000527629.1
ENST00000361723.3 ENST00000531408.1 |
FAM111A
|
family with sequence similarity 111, member A |
| chrX_+_139791917 | 0.02 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr1_+_209878182 | 0.02 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr17_-_48474828 | 0.02 |
ENST00000576448.1
ENST00000225972.7 |
LRRC59
|
leucine rich repeat containing 59 |
| chr8_-_141810634 | 0.02 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr12_-_10601963 | 0.02 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr12_+_19358228 | 0.02 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr3_-_69101413 | 0.02 |
ENST00000398559.2
|
TMF1
|
TATA element modulatory factor 1 |
| chrX_-_134936735 | 0.02 |
ENST00000487941.2
ENST00000434966.2 |
CT45A4
|
cancer/testis antigen family 45, member A4 |
| chr6_-_28973037 | 0.02 |
ENST00000377179.3
|
ZNF311
|
zinc finger protein 311 |
| chr6_+_125540951 | 0.02 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr2_+_87755054 | 0.02 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr8_-_91095099 | 0.02 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr19_+_45147098 | 0.02 |
ENST00000425690.3
ENST00000344956.4 ENST00000403059.4 |
PVR
|
poliovirus receptor |
| chr12_+_16064106 | 0.02 |
ENST00000428559.2
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr5_+_140501581 | 0.02 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr6_+_39760129 | 0.02 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr17_+_34312621 | 0.01 |
ENST00000591669.1
|
CTB-186H2.3
|
Uncharacterized protein |
| chr10_+_11207438 | 0.01 |
ENST00000609692.1
ENST00000354897.3 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr2_+_143635067 | 0.01 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chrX_-_30993201 | 0.01 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr19_+_55128576 | 0.01 |
ENST00000396331.1
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| chr7_+_21582638 | 0.01 |
ENST00000409508.3
ENST00000328843.6 |
DNAH11
|
dynein, axonemal, heavy chain 11 |
| chr4_-_1670632 | 0.01 |
ENST00000461064.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr15_+_40886199 | 0.01 |
ENST00000346991.5
ENST00000528975.1 ENST00000527044.1 |
CASC5
|
cancer susceptibility candidate 5 |
| chr19_+_44037546 | 0.01 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr17_-_60883993 | 0.01 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr5_+_80529104 | 0.01 |
ENST00000254035.4
ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr12_-_49245916 | 0.01 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr10_-_70287172 | 0.01 |
ENST00000539557.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chrX_-_134971059 | 0.01 |
ENST00000472834.1
|
CT45A6
|
cancer/testis antigen family 45, member A6 |
| chrX_+_134847185 | 0.01 |
ENST00000370741.3
ENST00000497301.2 |
CT45A1
|
cancer/testis antigen family 45, member A1 |
| chr13_-_33112956 | 0.01 |
ENST00000505213.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr1_+_111991474 | 0.01 |
ENST00000369722.3
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr4_+_86699834 | 0.01 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr19_-_13227463 | 0.01 |
ENST00000437766.1
ENST00000221504.8 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr5_+_75904950 | 0.01 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr12_-_123187890 | 0.01 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr19_+_11200038 | 0.01 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr12_+_8975061 | 0.01 |
ENST00000299698.7
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chrX_-_134971244 | 0.01 |
ENST00000491002.2
ENST00000448053.2 |
CT45A6
|
cancer/testis antigen family 45, member A6 |
| chrX_-_135338503 | 0.01 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr19_-_7812397 | 0.00 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr8_-_107782463 | 0.00 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chr3_+_44690211 | 0.00 |
ENST00000396056.2
ENST00000432115.2 ENST00000415571.2 ENST00000399560.2 ENST00000296092.3 ENST00000542250.1 ENST00000453164.1 |
ZNF35
|
zinc finger protein 35 |
| chr22_+_45725524 | 0.00 |
ENST00000405548.3
|
FAM118A
|
family with sequence similarity 118, member A |
| chr6_-_28303901 | 0.00 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr13_+_20207782 | 0.00 |
ENST00000414242.2
ENST00000361479.5 |
MPHOSPH8
|
M-phase phosphoprotein 8 |
| chr2_+_143635222 | 0.00 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr11_+_18230727 | 0.00 |
ENST00000527059.1
|
RP11-113D6.10
|
Putative mitochondrial carrier protein LOC494141 |
| chr12_-_12674032 | 0.00 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr7_-_99679324 | 0.00 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr14_-_100046444 | 0.00 |
ENST00000554996.1
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr11_-_104480019 | 0.00 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BCL6B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.3 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:1904440 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) negative regulation of natural killer cell activation(GO:0032815) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.1 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:2000843 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |