Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
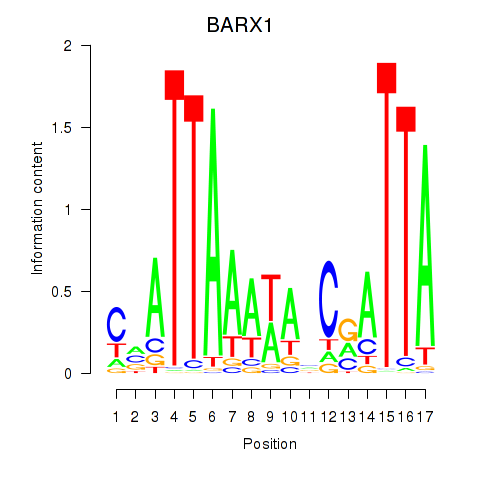
Results for BARX1
Z-value: 0.78
Transcription factors associated with BARX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARX1
|
ENSG00000131668.9 | BARX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BARX1 | hg19_v2_chr9_-_96717654_96717666 | 0.75 | 2.5e-01 | Click! |
Activity profile of BARX1 motif
Sorted Z-values of BARX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_29181503 | 0.76 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr11_-_9482010 | 0.58 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr7_-_34978980 | 0.50 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr6_+_26156551 | 0.50 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr13_+_113030658 | 0.44 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr6_+_27782788 | 0.37 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr22_-_32767017 | 0.36 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr12_-_10588539 | 0.36 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr5_+_150404904 | 0.35 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr3_-_172859017 | 0.35 |
ENST00000351008.3
|
SPATA16
|
spermatogenesis associated 16 |
| chr17_-_57158523 | 0.34 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr11_+_115498761 | 0.34 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr16_+_58074069 | 0.33 |
ENST00000570065.1
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr5_+_42756903 | 0.32 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr15_-_36544450 | 0.31 |
ENST00000561394.1
|
RP11-184D12.1
|
RP11-184D12.1 |
| chr19_-_14992264 | 0.30 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chrX_-_30871004 | 0.30 |
ENST00000378928.1
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr19_-_10491130 | 0.30 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr2_+_90198535 | 0.28 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr6_-_26285737 | 0.27 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr6_+_27775899 | 0.27 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr1_+_218683438 | 0.26 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr11_+_65266507 | 0.25 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr12_-_27235447 | 0.25 |
ENST00000429849.2
|
C12orf71
|
chromosome 12 open reading frame 71 |
| chr7_+_123469037 | 0.25 |
ENST00000489978.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr4_-_112993808 | 0.25 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr11_+_55606228 | 0.24 |
ENST00000378396.1
|
OR5D16
|
olfactory receptor, family 5, subfamily D, member 16 |
| chr19_-_53758094 | 0.24 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr13_-_49987885 | 0.23 |
ENST00000409082.1
|
CAB39L
|
calcium binding protein 39-like |
| chr19_-_53757941 | 0.23 |
ENST00000594517.1
ENST00000601413.1 ENST00000594681.1 |
ZNF677
|
zinc finger protein 677 |
| chr6_-_27782548 | 0.23 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr6_-_27775694 | 0.23 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr19_+_9361606 | 0.23 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr6_+_26273144 | 0.23 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr22_-_32766972 | 0.22 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr19_-_10491234 | 0.22 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr22_+_43011247 | 0.22 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr2_+_38177575 | 0.21 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr7_-_105221898 | 0.21 |
ENST00000486180.1
ENST00000485614.1 ENST00000480514.1 |
EFCAB10
|
EF-hand calcium binding domain 10 |
| chr19_+_52873166 | 0.21 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr15_-_72978490 | 0.21 |
ENST00000311755.3
|
HIGD2B
|
HIG1 hypoxia inducible domain family, member 2B |
| chr12_+_28605426 | 0.20 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chrX_-_55020511 | 0.20 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr4_+_74606223 | 0.20 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr16_+_28648975 | 0.20 |
ENST00000529716.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr8_-_112039643 | 0.19 |
ENST00000524283.1
|
RP11-946L20.2
|
RP11-946L20.2 |
| chr17_+_74729060 | 0.19 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chrX_+_69501943 | 0.19 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr2_+_242498135 | 0.19 |
ENST00000318407.3
|
BOK
|
BCL2-related ovarian killer |
| chr1_-_153514241 | 0.19 |
ENST00000368718.1
ENST00000359215.1 |
S100A5
|
S100 calcium binding protein A5 |
| chr14_+_77582905 | 0.19 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr10_-_74283694 | 0.18 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr2_-_14541060 | 0.18 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr11_+_114168085 | 0.18 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr10_-_79397740 | 0.18 |
ENST00000372440.1
ENST00000480683.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr5_+_169011033 | 0.18 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr6_+_26199737 | 0.18 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr6_+_123100853 | 0.18 |
ENST00000356535.4
|
FABP7
|
fatty acid binding protein 7, brain |
| chr16_+_28985542 | 0.18 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr4_-_38806404 | 0.18 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr6_+_31707725 | 0.18 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr9_+_133539981 | 0.17 |
ENST00000253008.2
|
PRDM12
|
PR domain containing 12 |
| chr5_-_56778635 | 0.17 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr6_+_118869452 | 0.17 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr16_-_28374829 | 0.17 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr10_-_5638048 | 0.17 |
ENST00000478294.1
|
RP13-463N16.6
|
RP13-463N16.6 |
| chr11_+_94300474 | 0.17 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr1_-_19615744 | 0.16 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr12_+_9822331 | 0.16 |
ENST00000545918.1
ENST00000543300.1 ENST00000261339.6 ENST00000466035.2 |
CLEC2D
|
C-type lectin domain family 2, member D |
| chr19_-_47349395 | 0.16 |
ENST00000597020.1
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr15_+_67813406 | 0.16 |
ENST00000342683.4
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr1_+_196743912 | 0.16 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr19_+_36142147 | 0.16 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr1_-_114429997 | 0.16 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr8_-_65730127 | 0.16 |
ENST00000522106.1
|
RP11-1D12.2
|
RP11-1D12.2 |
| chr4_-_69817481 | 0.16 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr4_+_11470867 | 0.15 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr12_+_110011571 | 0.15 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr13_+_108921977 | 0.15 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr19_-_56879416 | 0.14 |
ENST00000591036.1
|
AC006116.21
|
AC006116.21 |
| chr7_+_125078119 | 0.14 |
ENST00000458437.1
ENST00000415896.1 |
RP11-807H17.1
|
RP11-807H17.1 |
| chr7_+_141408153 | 0.14 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr13_-_49975632 | 0.14 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr19_+_3762645 | 0.14 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr22_+_51176624 | 0.14 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr2_-_236684438 | 0.14 |
ENST00000409661.1
|
AC064874.1
|
Uncharacterized protein |
| chr19_+_21579958 | 0.14 |
ENST00000339914.6
ENST00000599461.1 |
ZNF493
|
zinc finger protein 493 |
| chr3_-_180397256 | 0.14 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr22_-_19466732 | 0.14 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr11_-_114430570 | 0.14 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr12_-_58329819 | 0.13 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr1_+_43291220 | 0.13 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr22_-_50970919 | 0.13 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr9_-_95298254 | 0.13 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr2_-_190927447 | 0.13 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr4_+_69962212 | 0.13 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr3_-_196295437 | 0.13 |
ENST00000429115.1
|
WDR53
|
WD repeat domain 53 |
| chr12_-_57328187 | 0.13 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr7_+_115862858 | 0.13 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr2_+_3622932 | 0.13 |
ENST00000406376.1
|
RPS7
|
ribosomal protein S7 |
| chr19_+_13842559 | 0.13 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr4_-_90756769 | 0.13 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr14_+_73563735 | 0.12 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr7_+_13141010 | 0.12 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr5_+_140213815 | 0.12 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr3_-_197300194 | 0.12 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr4_+_69962185 | 0.12 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr6_+_37321823 | 0.12 |
ENST00000487950.1
ENST00000469731.1 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr5_+_140235469 | 0.12 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr4_+_39408470 | 0.12 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr3_-_149093499 | 0.12 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr8_-_18711866 | 0.12 |
ENST00000519851.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chrM_+_8366 | 0.12 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr14_+_38264418 | 0.12 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr4_+_174818390 | 0.12 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr4_-_88244010 | 0.12 |
ENST00000302219.6
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr15_+_75639372 | 0.12 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr1_+_59775752 | 0.12 |
ENST00000371212.1
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr19_-_23869970 | 0.12 |
ENST00000601010.1
|
ZNF675
|
zinc finger protein 675 |
| chr14_-_54425475 | 0.12 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr3_+_101659682 | 0.11 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr1_+_171283331 | 0.11 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr1_+_43424698 | 0.11 |
ENST00000431759.1
|
SLC2A1-AS1
|
SLC2A1 antisense RNA 1 |
| chr7_+_116595028 | 0.11 |
ENST00000397751.1
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr11_+_122709200 | 0.11 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr2_+_161993465 | 0.11 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr14_-_81425828 | 0.11 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr1_+_202091980 | 0.11 |
ENST00000367282.5
|
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr4_+_139694701 | 0.11 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chr8_-_101719159 | 0.11 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_-_49582978 | 0.11 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr15_-_55657428 | 0.11 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr4_+_177241094 | 0.11 |
ENST00000503362.1
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr4_-_88244049 | 0.10 |
ENST00000328546.4
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr15_+_90611465 | 0.10 |
ENST00000559360.1
|
ZNF710
|
zinc finger protein 710 |
| chr2_-_228497888 | 0.10 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr12_-_130529501 | 0.10 |
ENST00000561864.1
ENST00000567788.1 |
RP11-474D1.4
RP11-474D1.3
|
RP11-474D1.4 RP11-474D1.3 |
| chr3_-_196295385 | 0.10 |
ENST00000425888.1
|
WDR53
|
WD repeat domain 53 |
| chr11_-_22647350 | 0.10 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr13_+_25670268 | 0.10 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr2_+_130737223 | 0.10 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr22_-_35627045 | 0.10 |
ENST00000423311.1
|
CTA-714B7.5
|
CTA-714B7.5 |
| chr17_+_36908984 | 0.10 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr10_+_6625605 | 0.10 |
ENST00000414894.1
ENST00000449648.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr12_+_69186125 | 0.10 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr9_-_69229650 | 0.10 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr2_+_109403193 | 0.10 |
ENST00000412964.2
ENST00000295124.4 |
CCDC138
|
coiled-coil domain containing 138 |
| chr7_-_149158187 | 0.10 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr1_-_43855444 | 0.10 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr16_+_5121814 | 0.10 |
ENST00000262374.5
ENST00000586840.1 |
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr11_-_47546220 | 0.10 |
ENST00000528538.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr15_+_63050785 | 0.10 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr1_-_6420737 | 0.09 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr4_+_80584903 | 0.09 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr8_+_107460147 | 0.09 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chr17_-_42992856 | 0.09 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr9_-_95244781 | 0.09 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr6_+_28249299 | 0.09 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr11_-_77734260 | 0.09 |
ENST00000353172.5
|
KCTD14
|
potassium channel tetramerization domain containing 14 |
| chr12_+_96883347 | 0.09 |
ENST00000524981.4
ENST00000298953.3 |
C12orf55
|
chromosome 12 open reading frame 55 |
| chr19_-_48389651 | 0.09 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr1_+_196857144 | 0.09 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr17_-_73901494 | 0.09 |
ENST00000309352.3
|
MRPL38
|
mitochondrial ribosomal protein L38 |
| chr7_+_141463897 | 0.09 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr7_+_75508266 | 0.09 |
ENST00000006777.6
ENST00000318622.4 |
RHBDD2
|
rhomboid domain containing 2 |
| chr11_+_65154070 | 0.09 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr2_+_196313239 | 0.09 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr9_-_70465758 | 0.09 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr2_-_74007095 | 0.08 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr22_-_19466454 | 0.08 |
ENST00000494054.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr22_+_45148432 | 0.08 |
ENST00000389774.2
ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8
|
Rho GTPase activating protein 8 |
| chr7_+_64126503 | 0.08 |
ENST00000360117.3
|
ZNF107
|
zinc finger protein 107 |
| chr18_-_47813940 | 0.08 |
ENST00000586837.1
ENST00000412036.2 ENST00000589940.1 |
CXXC1
|
CXXC finger protein 1 |
| chr8_+_9046503 | 0.08 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr7_-_139763521 | 0.08 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr11_+_46958248 | 0.08 |
ENST00000536126.1
ENST00000278460.7 ENST00000378618.2 ENST00000395460.2 ENST00000378615.3 ENST00000543718.1 |
C11orf49
|
chromosome 11 open reading frame 49 |
| chr7_+_138818490 | 0.08 |
ENST00000430935.1
ENST00000495038.1 ENST00000474035.2 ENST00000478836.2 ENST00000464848.1 ENST00000343187.4 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr19_-_51611623 | 0.08 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr1_+_40974431 | 0.08 |
ENST00000296380.4
ENST00000432259.1 ENST00000418186.1 |
EXO5
|
exonuclease 5 |
| chr10_-_91403625 | 0.08 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr6_-_83775489 | 0.08 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr8_-_37707356 | 0.08 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr20_+_5987890 | 0.08 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr4_+_1385340 | 0.08 |
ENST00000324803.4
|
CRIPAK
|
cysteine-rich PAK1 inhibitor |
| chrX_+_41583408 | 0.08 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr13_+_24144509 | 0.08 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr17_-_47786375 | 0.08 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr15_+_75639296 | 0.08 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr5_+_159848807 | 0.08 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr3_-_196295468 | 0.08 |
ENST00000332629.5
ENST00000433160.1 |
WDR53
|
WD repeat domain 53 |
| chr22_-_19466683 | 0.08 |
ENST00000399523.1
ENST00000421968.2 ENST00000447868.1 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr11_+_118938485 | 0.08 |
ENST00000300793.6
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr8_+_1993173 | 0.08 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr1_+_16083154 | 0.08 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr10_-_1071796 | 0.08 |
ENST00000277517.1
|
IDI2
|
isopentenyl-diphosphate delta isomerase 2 |
| chr9_-_128246769 | 0.08 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr16_+_89724434 | 0.08 |
ENST00000568929.1
|
SPATA33
|
spermatogenesis associated 33 |
| chr12_+_94071341 | 0.08 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr12_-_54778244 | 0.07 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr7_-_137028498 | 0.07 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
Network of associatons between targets according to the STRING database.
First level regulatory network of BARX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.2 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.2 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 | 0.2 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.5 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.3 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0060503 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:1902164 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.0 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.0 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.5 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.3 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |