Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
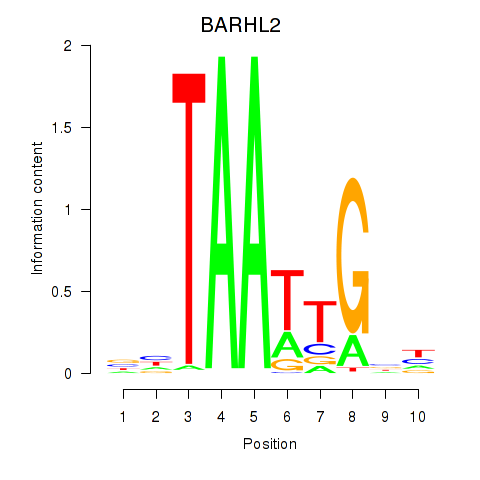
Results for BARHL2
Z-value: 0.43
Transcription factors associated with BARHL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARHL2
|
ENSG00000143032.7 | BarH like homeobox 2 |
Activity profile of BARHL2 motif
Sorted Z-values of BARHL2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94752349 | 0.35 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr1_-_168464875 | 0.34 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr17_+_61151306 | 0.33 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr4_+_76649753 | 0.32 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr13_+_110958124 | 0.24 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr8_-_25281747 | 0.21 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr2_-_87248975 | 0.20 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr14_-_91294472 | 0.17 |
ENST00000555975.1
|
CTD-3035D6.2
|
CTD-3035D6.2 |
| chr17_+_76037081 | 0.16 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr8_+_19536083 | 0.15 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr1_-_1208851 | 0.15 |
ENST00000488418.1
|
UBE2J2
|
ubiquitin-conjugating enzyme E2, J2 |
| chr2_-_203735586 | 0.14 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr12_-_76879852 | 0.14 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr17_-_46799872 | 0.12 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr4_+_147096837 | 0.12 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_+_32666188 | 0.12 |
ENST00000421922.2
|
CCDC28B
|
coiled-coil domain containing 28B |
| chrX_-_49056635 | 0.12 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr1_+_225600404 | 0.11 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr14_-_107283278 | 0.11 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr15_-_37393406 | 0.11 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr7_-_84121858 | 0.11 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr18_+_5238549 | 0.11 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr12_-_94673956 | 0.11 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr4_+_129732419 | 0.10 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr8_+_32579321 | 0.10 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr12_-_91546926 | 0.10 |
ENST00000550758.1
|
DCN
|
decorin |
| chr12_-_120241187 | 0.10 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chrX_+_139791917 | 0.10 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr9_-_98079154 | 0.10 |
ENST00000433829.1
|
FANCC
|
Fanconi anemia, complementation group C |
| chr1_-_12908578 | 0.10 |
ENST00000317869.6
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C-like 1 |
| chr11_+_110001723 | 0.10 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chrX_+_78003204 | 0.09 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr11_-_33913708 | 0.09 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr5_+_135496675 | 0.09 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr8_-_150563 | 0.09 |
ENST00000523795.2
|
RP11-585F1.10
|
Protein LOC100286914 |
| chr11_-_118023594 | 0.09 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr17_-_38928414 | 0.09 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr12_-_86650077 | 0.09 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_-_1922789 | 0.09 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr8_-_97247759 | 0.09 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr15_+_76016293 | 0.09 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr21_+_17792672 | 0.09 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_-_124553437 | 0.09 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr8_+_22424551 | 0.09 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr14_+_35514323 | 0.09 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr6_+_144665237 | 0.08 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr21_-_19858196 | 0.08 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr4_-_120243545 | 0.08 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr4_+_155484103 | 0.08 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr3_-_107777208 | 0.08 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr7_-_86849836 | 0.08 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr6_-_76203345 | 0.08 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_+_70916119 | 0.08 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr14_+_61449197 | 0.08 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr9_+_109685630 | 0.08 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr3_-_120400960 | 0.08 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr17_-_7307358 | 0.08 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr12_-_91573132 | 0.08 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr6_-_99873145 | 0.08 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr12_+_14572070 | 0.07 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr7_-_111032971 | 0.07 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_7014126 | 0.07 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr14_+_20187174 | 0.07 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chrX_+_10126488 | 0.07 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr16_-_80926457 | 0.07 |
ENST00000563626.1
ENST00000562231.1 |
RP11-314O13.1
|
RP11-314O13.1 |
| chr3_+_98699880 | 0.07 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr15_+_58702742 | 0.07 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr8_+_32579271 | 0.07 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr18_+_5238055 | 0.07 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr3_-_196910477 | 0.07 |
ENST00000447466.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr4_-_140527848 | 0.07 |
ENST00000608795.1
ENST00000608958.1 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr6_+_131571535 | 0.07 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr17_-_55162360 | 0.07 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr12_-_54652060 | 0.07 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr19_-_41870026 | 0.07 |
ENST00000243578.3
|
B9D2
|
B9 protein domain 2 |
| chr18_-_33702078 | 0.07 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr13_+_111855399 | 0.07 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr5_-_78281623 | 0.07 |
ENST00000521117.1
|
ARSB
|
arylsulfatase B |
| chr1_+_84629976 | 0.07 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr16_+_53133070 | 0.07 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_130160440 | 0.06 |
ENST00000439597.1
ENST00000423934.1 |
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr17_+_67498538 | 0.06 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr22_+_46476192 | 0.06 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chrX_-_55057403 | 0.06 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr11_-_111649074 | 0.06 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr2_-_208031943 | 0.06 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr5_+_108083517 | 0.06 |
ENST00000281092.4
ENST00000536402.1 |
FER
|
fer (fps/fes related) tyrosine kinase |
| chr6_-_32157947 | 0.06 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr6_-_26189304 | 0.06 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr16_+_14280742 | 0.06 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chrX_+_86772787 | 0.06 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chrX_-_71351678 | 0.06 |
ENST00000609883.1
ENST00000545866.1 |
RGAG4
|
retrotransposon gag domain containing 4 |
| chr11_-_72070206 | 0.06 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr3_-_9834463 | 0.06 |
ENST00000439043.1
|
TADA3
|
transcriptional adaptor 3 |
| chr4_+_169418255 | 0.06 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr8_-_125486755 | 0.06 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr10_-_49860525 | 0.06 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr10_-_75351088 | 0.06 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr9_+_131703757 | 0.06 |
ENST00000482796.1
|
RP11-101E3.5
|
RP11-101E3.5 |
| chr11_+_72983246 | 0.06 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr11_-_111649015 | 0.06 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr5_-_13944652 | 0.06 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr12_+_54694979 | 0.06 |
ENST00000552848.1
|
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr18_+_29171689 | 0.06 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr5_+_150639360 | 0.05 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr10_-_97200772 | 0.05 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr2_+_120189422 | 0.05 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr6_-_76203454 | 0.05 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr10_+_135050908 | 0.05 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr1_+_196788887 | 0.05 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr5_-_146781153 | 0.05 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr8_+_32579341 | 0.05 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr11_+_62496114 | 0.05 |
ENST00000532583.1
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chrX_-_16730688 | 0.05 |
ENST00000359276.4
|
CTPS2
|
CTP synthase 2 |
| chr8_-_128231299 | 0.05 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr11_+_10772847 | 0.05 |
ENST00000524523.1
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr1_+_145439306 | 0.05 |
ENST00000425134.1
|
TXNIP
|
thioredoxin interacting protein |
| chr5_-_133510456 | 0.05 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr1_-_78444738 | 0.05 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr2_-_70944855 | 0.05 |
ENST00000415348.1
|
ADD2
|
adducin 2 (beta) |
| chr21_+_43619796 | 0.05 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr1_-_78444776 | 0.05 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr12_+_52056548 | 0.05 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr2_-_73869508 | 0.05 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr5_+_140855495 | 0.05 |
ENST00000308177.3
|
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr7_+_99717230 | 0.05 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr7_+_80275953 | 0.05 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_-_71551868 | 0.05 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr22_-_36013368 | 0.05 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr15_+_63682335 | 0.05 |
ENST00000559379.1
ENST00000559821.1 |
RP11-321G12.1
|
RP11-321G12.1 |
| chr4_-_76649546 | 0.05 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr12_-_75784669 | 0.05 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr7_+_34697973 | 0.05 |
ENST00000381539.3
|
NPSR1
|
neuropeptide S receptor 1 |
| chr2_+_66662249 | 0.05 |
ENST00000560281.2
|
MEIS1
|
Meis homeobox 1 |
| chr2_+_149402989 | 0.05 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chrX_+_86772707 | 0.05 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr11_+_64018955 | 0.05 |
ENST00000279230.6
ENST00000540288.1 ENST00000325234.5 |
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr9_+_2157655 | 0.05 |
ENST00000452193.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_224467002 | 0.05 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr6_+_26087509 | 0.05 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr16_+_69345243 | 0.05 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr3_+_138340049 | 0.05 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr9_+_6215799 | 0.04 |
ENST00000417746.2
ENST00000456383.2 |
IL33
|
interleukin 33 |
| chr2_+_48796120 | 0.04 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr8_+_86157699 | 0.04 |
ENST00000321764.3
|
CA13
|
carbonic anhydrase XIII |
| chr14_+_105331596 | 0.04 |
ENST00000556508.1
ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B
|
centrosomal protein 170B |
| chr14_+_73706308 | 0.04 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr8_+_82066514 | 0.04 |
ENST00000519412.1
ENST00000521953.1 |
RP11-1149M10.2
|
RP11-1149M10.2 |
| chr3_+_193965426 | 0.04 |
ENST00000455821.1
|
RP11-513G11.3
|
RP11-513G11.3 |
| chr13_+_95364963 | 0.04 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr9_-_134955246 | 0.04 |
ENST00000357028.2
ENST00000474263.1 ENST00000292035.5 |
MED27
|
mediator complex subunit 27 |
| chr12_+_14561422 | 0.04 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr12_+_76653682 | 0.04 |
ENST00000553247.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr5_+_31193847 | 0.04 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chrX_+_49832231 | 0.04 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr10_+_7745303 | 0.04 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr13_-_95364389 | 0.04 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr11_+_114310164 | 0.04 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr13_+_111766897 | 0.04 |
ENST00000317133.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr14_-_91720224 | 0.04 |
ENST00000238699.3
ENST00000531499.2 |
GPR68
|
G protein-coupled receptor 68 |
| chr1_+_92545862 | 0.04 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr4_+_56814968 | 0.04 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr5_-_111312622 | 0.04 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr4_+_86748898 | 0.04 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr12_-_15815626 | 0.04 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr12_-_118628350 | 0.04 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr6_-_116833500 | 0.04 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr7_-_27169801 | 0.04 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr1_+_236958554 | 0.04 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr11_-_64684672 | 0.04 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr2_-_220110111 | 0.04 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr18_-_5238525 | 0.04 |
ENST00000581170.1
ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5
LINC00526
|
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr2_+_152214098 | 0.04 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr1_-_93257951 | 0.04 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr7_+_13141010 | 0.04 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr3_+_150126101 | 0.04 |
ENST00000361875.3
ENST00000361136.2 |
TSC22D2
|
TSC22 domain family, member 2 |
| chr5_+_140710061 | 0.04 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr12_-_118628315 | 0.04 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr1_-_243326612 | 0.04 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr3_-_138048682 | 0.04 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr6_+_20534672 | 0.04 |
ENST00000274695.4
ENST00000378624.4 |
CDKAL1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr2_-_231989808 | 0.04 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr9_-_7800067 | 0.04 |
ENST00000358227.4
|
TMEM261
|
transmembrane protein 261 |
| chr16_-_2031464 | 0.04 |
ENST00000356120.4
ENST00000354249.4 |
NOXO1
|
NADPH oxidase organizer 1 |
| chr4_-_119759795 | 0.04 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr1_+_84630352 | 0.04 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_+_126315091 | 0.04 |
ENST00000335110.5
|
FAT4
|
FAT atypical cadherin 4 |
| chr2_+_135596180 | 0.04 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr3_-_123339343 | 0.04 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr1_+_179263308 | 0.04 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr9_-_5833027 | 0.04 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr1_+_236558694 | 0.04 |
ENST00000359362.5
|
EDARADD
|
EDAR-associated death domain |
| chrX_-_16730984 | 0.04 |
ENST00000380241.3
|
CTPS2
|
CTP synthase 2 |
| chr1_-_101360205 | 0.04 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr19_+_984313 | 0.04 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chr16_+_2031633 | 0.04 |
ENST00000598236.1
|
AC005606.1
|
LOC283868 protein; Uncharacterized protein |
| chr2_-_71454185 | 0.04 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr6_-_134373732 | 0.04 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr18_+_21529811 | 0.04 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr7_-_84122033 | 0.04 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
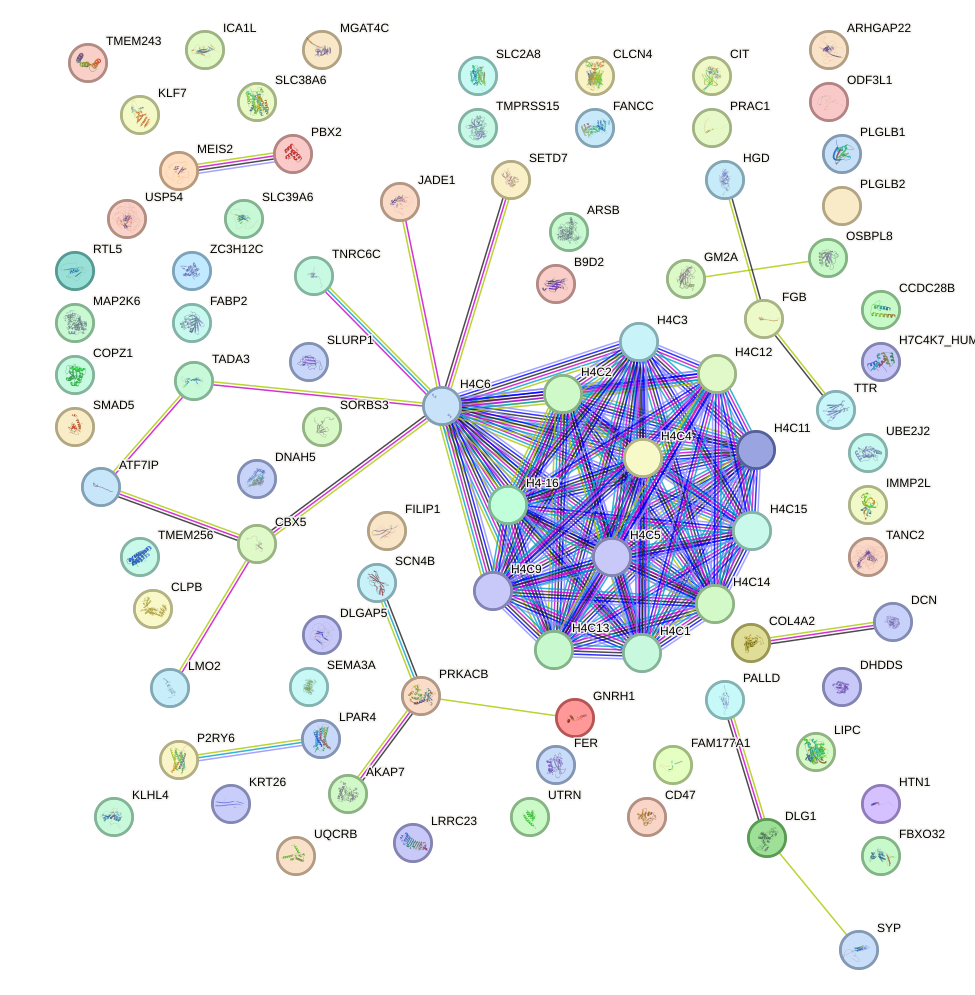
Network of associatons between targets according to the STRING database.
First level regulatory network of BARHL2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0050904 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0061045 | negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.0 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.0 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.0 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |