Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
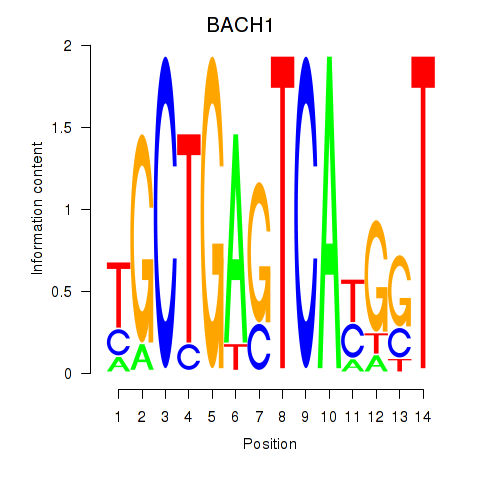
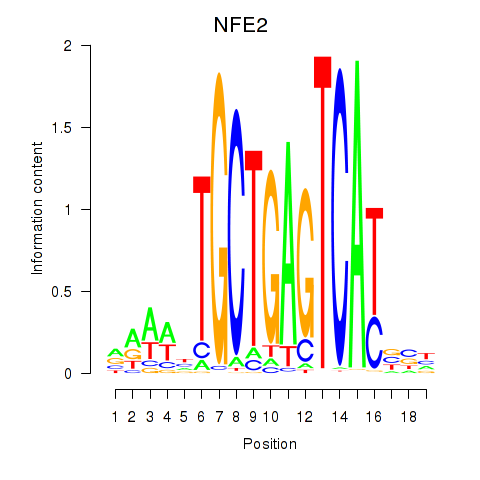
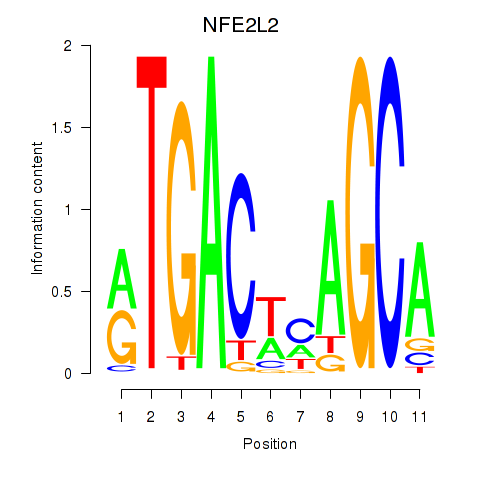
Results for BACH1_NFE2_NFE2L2
Z-value: 0.52
Transcription factors associated with BACH1_NFE2_NFE2L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH1
|
ENSG00000156273.11 | BTB domain and CNC homolog 1 |
|
NFE2
|
ENSG00000123405.9 | nuclear factor, erythroid 2 |
|
NFE2L2
|
ENSG00000116044.11 | nuclear factor, erythroid 2 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L2 | hg19_v2_chr2_-_178128528_178128574 | -0.98 | 1.5e-02 | Click! |
| BACH1 | hg19_v2_chr21_+_30672433_30672464 | -0.57 | 4.3e-01 | Click! |
| NFE2 | hg19_v2_chr12_-_54689532_54689551 | -0.01 | 9.9e-01 | Click! |
Activity profile of BACH1_NFE2_NFE2L2 motif
Sorted Z-values of BACH1_NFE2_NFE2L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_67141640 | 0.46 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr22_+_45072925 | 0.38 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr19_+_18699535 | 0.34 |
ENST00000358607.6
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr22_+_45072958 | 0.34 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr14_+_71165292 | 0.28 |
ENST00000553682.1
|
RP6-65G23.1
|
RP6-65G23.1 |
| chr11_+_67351213 | 0.27 |
ENST00000398603.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr16_+_83986827 | 0.25 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr19_-_10613361 | 0.21 |
ENST00000591039.1
ENST00000591419.1 |
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr10_-_75532373 | 0.21 |
ENST00000595757.1
|
AC022400.2
|
Uncharacterized protein; cDNA FLJ44715 fis, clone BRACE3021430 |
| chr10_-_76995769 | 0.19 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr22_-_39268192 | 0.19 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr16_-_1429627 | 0.19 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr4_+_22999152 | 0.19 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr8_+_125283924 | 0.18 |
ENST00000517482.1
|
RP11-383J24.1
|
RP11-383J24.1 |
| chr5_-_176923803 | 0.18 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr16_+_57662527 | 0.18 |
ENST00000563374.1
ENST00000568234.1 ENST00000565770.1 ENST00000564338.1 ENST00000566164.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr9_-_116840728 | 0.18 |
ENST00000265132.3
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr17_-_79881408 | 0.17 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr12_+_10365082 | 0.16 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr17_+_80317121 | 0.16 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr6_+_33378517 | 0.16 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr5_+_179247759 | 0.16 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr6_-_28321971 | 0.15 |
ENST00000396838.2
ENST00000426434.1 ENST00000434036.1 ENST00000439628.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr17_-_61512545 | 0.15 |
ENST00000585153.1
|
CYB561
|
cytochrome b561 |
| chr14_-_23504087 | 0.15 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr10_-_76995675 | 0.15 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr12_+_10366016 | 0.15 |
ENST00000546017.1
ENST00000535576.1 ENST00000539170.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr19_-_10613421 | 0.15 |
ENST00000393623.2
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr16_+_57662596 | 0.15 |
ENST00000567397.1
ENST00000568979.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_+_30418910 | 0.14 |
ENST00000566625.1
|
ZNF771
|
zinc finger protein 771 |
| chr5_+_35852797 | 0.14 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr19_+_18699599 | 0.14 |
ENST00000450195.2
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr11_+_67351019 | 0.14 |
ENST00000398606.3
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr10_-_71892555 | 0.14 |
ENST00000307864.1
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr9_+_140083099 | 0.14 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr8_+_94752349 | 0.13 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr16_-_1429674 | 0.13 |
ENST00000403703.1
ENST00000397464.1 ENST00000402641.2 |
UNKL
|
unkempt family zinc finger-like |
| chr9_-_140082983 | 0.13 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr19_-_10613862 | 0.13 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr2_-_220117867 | 0.13 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr7_-_86849836 | 0.12 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr1_-_158656488 | 0.12 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr3_+_14474178 | 0.12 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr1_+_76251912 | 0.12 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr16_+_12059091 | 0.12 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr11_+_60699222 | 0.11 |
ENST00000536409.1
|
TMEM132A
|
transmembrane protein 132A |
| chr2_-_220083076 | 0.11 |
ENST00000295750.4
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr8_-_94753202 | 0.11 |
ENST00000521947.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr22_+_29702572 | 0.11 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr5_-_176923846 | 0.11 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr17_+_7123207 | 0.11 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr17_+_21729593 | 0.11 |
ENST00000581769.1
ENST00000584755.1 |
UBBP4
|
ubiquitin B pseudogene 4 |
| chr22_+_18893736 | 0.10 |
ENST00000331444.6
|
DGCR6
|
DiGeorge syndrome critical region gene 6 |
| chr17_+_21729899 | 0.10 |
ENST00000583708.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr16_+_47496023 | 0.10 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr7_-_110174754 | 0.10 |
ENST00000435466.1
|
AC003088.1
|
AC003088.1 |
| chr10_-_71906342 | 0.10 |
ENST00000287078.6
ENST00000335494.5 |
TYSND1
|
trypsin domain containing 1 |
| chr6_-_33285505 | 0.10 |
ENST00000431845.2
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr5_+_98264867 | 0.10 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr8_-_50466973 | 0.10 |
ENST00000520800.1
|
RP11-738G5.2
|
Uncharacterized protein |
| chr6_+_33378738 | 0.10 |
ENST00000374512.3
ENST00000374516.3 |
PHF1
|
PHD finger protein 1 |
| chr1_+_31885963 | 0.10 |
ENST00000373709.3
|
SERINC2
|
serine incorporator 2 |
| chr22_+_18893816 | 0.10 |
ENST00000608842.1
|
DGCR6
|
DiGeorge syndrome critical region gene 6 |
| chr12_+_6644443 | 0.10 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr12_-_58026426 | 0.10 |
ENST00000418555.2
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr20_-_634000 | 0.10 |
ENST00000381962.3
|
SRXN1
|
sulfiredoxin 1 |
| chr2_+_54342533 | 0.10 |
ENST00000406041.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr11_+_72983246 | 0.09 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr8_+_123875624 | 0.09 |
ENST00000534247.1
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr11_-_62380199 | 0.09 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr12_+_10365404 | 0.09 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr7_-_35734607 | 0.09 |
ENST00000427455.1
|
HERPUD2
|
HERPUD family member 2 |
| chr12_-_122238913 | 0.09 |
ENST00000537157.1
|
AC084018.1
|
AC084018.1 |
| chr21_+_33671264 | 0.09 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr12_-_57030096 | 0.09 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr19_+_6531010 | 0.09 |
ENST00000245817.3
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9 |
| chr11_-_62474803 | 0.08 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_-_3862206 | 0.08 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr19_+_50094866 | 0.08 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr12_+_56075330 | 0.08 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr6_-_39902185 | 0.08 |
ENST00000373195.3
ENST00000308559.7 ENST00000373188.2 |
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr19_-_19739321 | 0.08 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr19_+_34850385 | 0.08 |
ENST00000587521.2
ENST00000587384.1 ENST00000592277.1 |
GPI
|
glucose-6-phosphate isomerase |
| chr17_-_3794021 | 0.08 |
ENST00000381769.2
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr19_+_18496957 | 0.08 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr16_+_57662419 | 0.08 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr10_-_76868931 | 0.08 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr5_-_175964366 | 0.08 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr14_-_65409502 | 0.08 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr2_+_201987200 | 0.08 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_120189422 | 0.08 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr16_+_67261008 | 0.08 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr18_+_3450161 | 0.07 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr16_+_57662138 | 0.07 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_-_67260901 | 0.07 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr8_-_145013711 | 0.07 |
ENST00000345136.3
|
PLEC
|
plectin |
| chr8_-_145025044 | 0.07 |
ENST00000322810.4
|
PLEC
|
plectin |
| chr19_+_56652643 | 0.07 |
ENST00000586123.1
|
ZNF444
|
zinc finger protein 444 |
| chr12_-_58026451 | 0.07 |
ENST00000552350.1
ENST00000548888.1 |
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr4_-_987217 | 0.07 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr12_+_104680793 | 0.07 |
ENST00000529546.1
ENST00000529751.1 ENST00000540716.1 ENST00000528079.2 ENST00000526580.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr14_+_103589789 | 0.07 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr7_+_100026406 | 0.07 |
ENST00000414441.1
|
MEPCE
|
methylphosphate capping enzyme |
| chr11_+_65339820 | 0.07 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.1 |
FAM89B
|
family with sequence similarity 89, member B |
| chr5_+_150020214 | 0.07 |
ENST00000307662.4
|
SYNPO
|
synaptopodin |
| chr7_+_1570322 | 0.07 |
ENST00000343242.4
|
MAFK
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr12_+_104680659 | 0.07 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr19_-_10444188 | 0.07 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chrX_+_153769446 | 0.07 |
ENST00000422680.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr11_-_66103867 | 0.06 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr16_+_30953696 | 0.06 |
ENST00000566320.2
ENST00000565939.1 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr8_-_142318398 | 0.06 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr1_-_156399184 | 0.06 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr1_-_150208291 | 0.06 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr22_+_42196666 | 0.06 |
ENST00000402061.3
ENST00000255784.5 |
CCDC134
|
coiled-coil domain containing 134 |
| chr10_+_75532028 | 0.06 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chrX_-_153718953 | 0.06 |
ENST00000369649.4
ENST00000393586.1 |
SLC10A3
|
solute carrier family 10, member 3 |
| chr3_-_48601206 | 0.06 |
ENST00000273610.3
|
UCN2
|
urocortin 2 |
| chr11_-_2924720 | 0.06 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr1_+_36554470 | 0.06 |
ENST00000373178.4
|
ADPRHL2
|
ADP-ribosylhydrolase like 2 |
| chr10_+_106014468 | 0.06 |
ENST00000369710.4
ENST00000369713.5 ENST00000445155.1 |
GSTO1
|
glutathione S-transferase omega 1 |
| chr6_-_33266492 | 0.06 |
ENST00000425946.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr9_+_35538616 | 0.06 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr16_+_29819446 | 0.06 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr8_-_94753184 | 0.06 |
ENST00000520560.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr2_-_11606275 | 0.06 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr1_-_12677714 | 0.06 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr17_+_18625336 | 0.06 |
ENST00000395671.4
ENST00000571542.1 ENST00000395672.2 ENST00000414850.2 ENST00000424146.2 |
TRIM16L
|
tripartite motif containing 16-like |
| chr22_+_20008646 | 0.06 |
ENST00000401833.1
|
TANGO2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr8_+_87111059 | 0.06 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr19_-_46142637 | 0.06 |
ENST00000590043.1
ENST00000589876.1 |
EML2
|
echinoderm microtubule associated protein like 2 |
| chrX_+_99899180 | 0.06 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chrX_+_153769409 | 0.06 |
ENST00000440286.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr16_-_67260691 | 0.06 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr18_+_21594384 | 0.06 |
ENST00000584250.1
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr9_+_116917807 | 0.05 |
ENST00000356083.3
|
COL27A1
|
collagen, type XXVII, alpha 1 |
| chr15_+_41851211 | 0.05 |
ENST00000263798.3
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr5_+_150020240 | 0.05 |
ENST00000519664.1
|
SYNPO
|
synaptopodin |
| chr17_+_16284399 | 0.05 |
ENST00000535788.1
|
UBB
|
ubiquitin B |
| chr15_-_83240507 | 0.05 |
ENST00000564522.1
ENST00000398592.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr6_-_24721054 | 0.05 |
ENST00000378119.4
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr16_+_56642041 | 0.05 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr3_+_100053625 | 0.05 |
ENST00000497785.1
|
NIT2
|
nitrilase family, member 2 |
| chr17_+_41476327 | 0.05 |
ENST00000320033.4
|
ARL4D
|
ADP-ribosylation factor-like 4D |
| chr11_-_128894053 | 0.05 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_-_170588163 | 0.05 |
ENST00000295830.8
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr17_-_30470154 | 0.05 |
ENST00000398832.2
|
AC090616.2
|
Uncharacterized protein |
| chr8_-_81787006 | 0.05 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr17_-_55162360 | 0.05 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr11_-_119066545 | 0.05 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr16_-_11036300 | 0.05 |
ENST00000331808.4
|
DEXI
|
Dexi homolog (mouse) |
| chr10_-_99531709 | 0.05 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr16_-_69760409 | 0.05 |
ENST00000561500.1
ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr22_-_32058166 | 0.05 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr22_-_19435755 | 0.05 |
ENST00000542103.1
ENST00000399562.4 |
C22orf39
|
chromosome 22 open reading frame 39 |
| chr17_+_75372165 | 0.05 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chr19_-_39390440 | 0.05 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr10_-_127505167 | 0.05 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr22_-_32058416 | 0.05 |
ENST00000439502.2
|
PISD
|
phosphatidylserine decarboxylase |
| chr19_+_56652686 | 0.05 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr12_-_49523896 | 0.05 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr2_-_190044480 | 0.05 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr1_-_200992827 | 0.05 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr22_-_45608324 | 0.05 |
ENST00000496226.1
ENST00000251993.7 |
KIAA0930
|
KIAA0930 |
| chr10_-_21786179 | 0.05 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr11_+_35222629 | 0.05 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr16_+_30077098 | 0.05 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr19_-_16045619 | 0.05 |
ENST00000402119.4
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr3_+_120315160 | 0.05 |
ENST00000485064.1
ENST00000492739.1 |
NDUFB4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
| chr11_-_66103932 | 0.04 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr10_-_73611046 | 0.04 |
ENST00000394934.1
ENST00000394936.3 |
PSAP
|
prosaposin |
| chr6_+_32121218 | 0.04 |
ENST00000414204.1
ENST00000361568.2 ENST00000395523.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr7_+_73275483 | 0.04 |
ENST00000320531.2
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28 |
| chr1_-_150208320 | 0.04 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_-_120687948 | 0.04 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr2_+_29353520 | 0.04 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr3_-_149293990 | 0.04 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr15_+_65914260 | 0.04 |
ENST00000261892.6
ENST00000339868.6 |
SLC24A1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr2_+_74781828 | 0.04 |
ENST00000340004.6
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr3_+_100053542 | 0.04 |
ENST00000394140.4
|
NIT2
|
nitrilase family, member 2 |
| chr19_-_39390350 | 0.04 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr9_+_141107506 | 0.04 |
ENST00000446912.2
|
FAM157B
|
family with sequence similarity 157, member B |
| chr9_-_34458531 | 0.04 |
ENST00000379089.1
ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A
|
family with sequence similarity 219, member A |
| chr7_+_1084206 | 0.04 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr5_-_93954227 | 0.04 |
ENST00000513200.3
ENST00000329378.7 ENST00000312498.7 |
KIAA0825
|
KIAA0825 |
| chr20_+_60878005 | 0.04 |
ENST00000253003.2
|
ADRM1
|
adhesion regulating molecule 1 |
| chr17_-_79604075 | 0.04 |
ENST00000374747.5
ENST00000539314.1 ENST00000331134.6 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr12_-_57030115 | 0.04 |
ENST00000379441.3
ENST00000179765.5 ENST00000551812.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr11_-_124981475 | 0.04 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr15_-_43212836 | 0.04 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr9_-_127533519 | 0.04 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr22_-_40859415 | 0.04 |
ENST00000402630.1
ENST00000407029.1 |
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr11_-_14541872 | 0.04 |
ENST00000419365.2
ENST00000530457.1 ENST00000532256.1 ENST00000533068.1 |
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1 |
| chr12_-_125401885 | 0.04 |
ENST00000542416.1
|
UBC
|
ubiquitin C |
| chr9_-_139922631 | 0.04 |
ENST00000341511.6
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr6_-_28321909 | 0.04 |
ENST00000446222.1
|
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr4_-_101111615 | 0.04 |
ENST00000273990.2
|
DDIT4L
|
DNA-damage-inducible transcript 4-like |
| chr7_+_95401877 | 0.04 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr2_+_54342574 | 0.04 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr19_-_46142680 | 0.04 |
ENST00000245925.3
|
EML2
|
echinoderm microtubule associated protein like 2 |
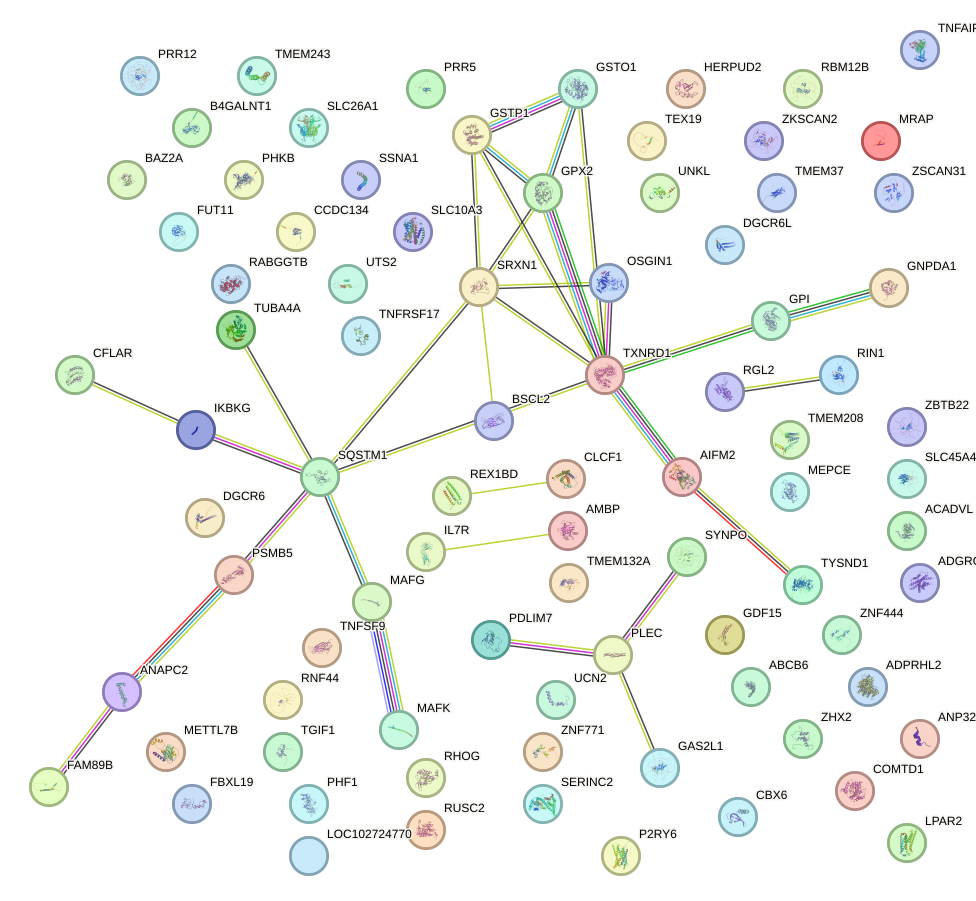
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH1_NFE2_NFE2L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.5 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.0 | GO:0051150 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0012502 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.0 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) corticotropin hormone receptor binding(GO:0031780) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.2 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.0 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |