Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
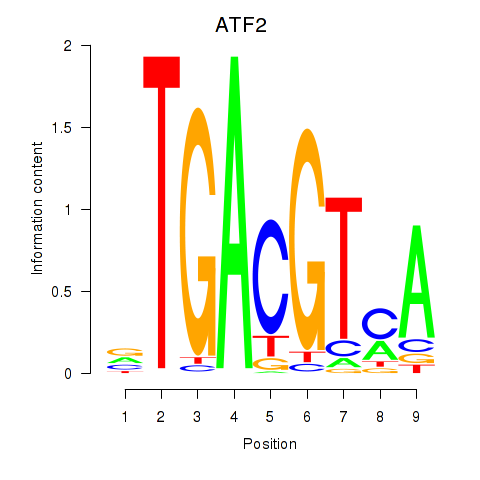
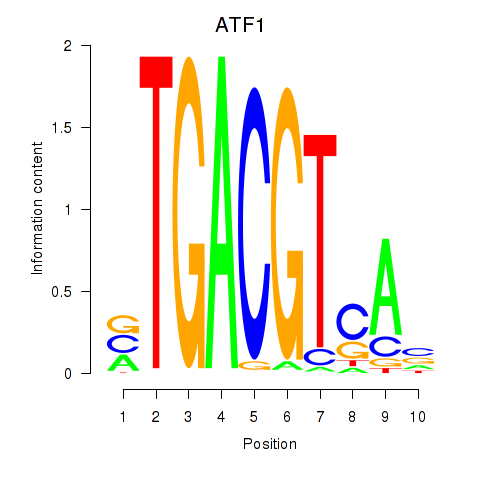
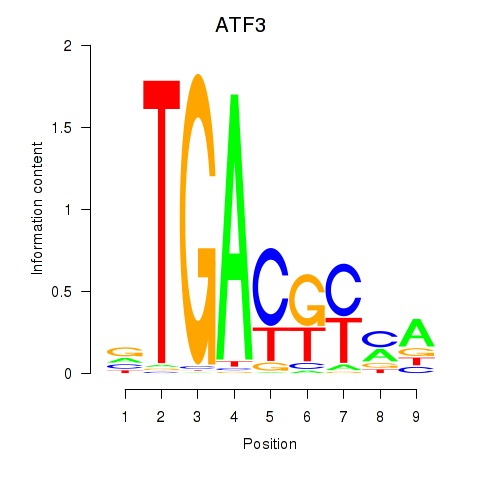
Results for ATF2_ATF1_ATF3
Z-value: 0.87
Transcription factors associated with ATF2_ATF1_ATF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF2
|
ENSG00000115966.12 | activating transcription factor 2 |
|
ATF1
|
ENSG00000123268.4 | activating transcription factor 1 |
|
ATF3
|
ENSG00000162772.12 | activating transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF3 | hg19_v2_chr1_+_212738676_212738755 | -0.84 | 1.6e-01 | Click! |
| ATF2 | hg19_v2_chr2_-_176032843_176032941 | -0.81 | 1.9e-01 | Click! |
| ATF1 | hg19_v2_chr12_+_51158263_51158395 | -0.73 | 2.7e-01 | Click! |
Activity profile of ATF2_ATF1_ATF3 motif
Sorted Z-values of ATF2_ATF1_ATF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_50979981 | 1.29 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr20_+_33146510 | 0.76 |
ENST00000397709.1
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr16_+_29911666 | 0.71 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr22_+_24236191 | 0.66 |
ENST00000215754.7
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr16_+_3068393 | 0.65 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr2_-_220408260 | 0.64 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr19_-_51014345 | 0.62 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr19_-_51014460 | 0.62 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr2_-_220408430 | 0.60 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr19_-_51014588 | 0.58 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr8_-_103876340 | 0.58 |
ENST00000518353.1
|
AZIN1
|
antizyme inhibitor 1 |
| chr11_+_63742050 | 0.58 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr16_+_29911864 | 0.58 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr7_-_105926058 | 0.54 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr5_+_61708488 | 0.53 |
ENST00000505902.1
|
IPO11
|
importin 11 |
| chr11_+_66610883 | 0.52 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr17_-_8059638 | 0.52 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr8_-_21966893 | 0.50 |
ENST00000522405.1
ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr11_-_9482010 | 0.50 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr11_+_66059339 | 0.48 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr19_+_50979753 | 0.47 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr19_+_55996316 | 0.46 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr7_-_65958525 | 0.46 |
ENST00000450353.1
ENST00000412091.1 |
GS1-124K5.3
|
GS1-124K5.3 |
| chr1_+_156698743 | 0.45 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chrX_+_153237740 | 0.44 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chr14_-_102976135 | 0.43 |
ENST00000560748.1
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr8_+_17104401 | 0.43 |
ENST00000324815.3
ENST00000518038.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr9_-_130477912 | 0.42 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr6_+_30029008 | 0.39 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr6_-_31940065 | 0.39 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr12_+_10365082 | 0.38 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr17_-_43210580 | 0.38 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr11_-_66056596 | 0.38 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr16_+_67694849 | 0.37 |
ENST00000602551.1
ENST00000458121.2 ENST00000219255.3 |
PARD6A
|
par-6 family cell polarity regulator alpha |
| chr17_+_685513 | 0.36 |
ENST00000304478.4
|
RNMTL1
|
RNA methyltransferase like 1 |
| chr19_-_18392422 | 0.36 |
ENST00000252818.3
|
JUND
|
jun D proto-oncogene |
| chr19_-_47734448 | 0.36 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr16_+_2563871 | 0.36 |
ENST00000330398.4
ENST00000568562.1 ENST00000569317.1 |
ATP6V0C
ATP6C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c Uncharacterized protein |
| chr16_+_2564254 | 0.36 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr19_-_47616992 | 0.35 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr12_+_51632638 | 0.35 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr3_+_184053703 | 0.35 |
ENST00000450976.1
ENST00000418281.1 ENST00000340957.5 ENST00000433578.1 |
FAM131A
|
family with sequence similarity 131, member A |
| chr19_-_54974894 | 0.34 |
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9 |
| chr17_-_36831156 | 0.34 |
ENST00000325814.5
|
C17orf96
|
chromosome 17 open reading frame 96 |
| chr10_+_134210672 | 0.34 |
ENST00000305233.5
ENST00000368609.4 |
PWWP2B
|
PWWP domain containing 2B |
| chr19_-_42759300 | 0.34 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr6_-_34664612 | 0.34 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr19_+_1941117 | 0.34 |
ENST00000255641.8
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr14_+_105219437 | 0.34 |
ENST00000329967.6
ENST00000347067.5 ENST00000553810.1 |
SIVA1
|
SIVA1, apoptosis-inducing factor |
| chr14_-_77495007 | 0.33 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr14_-_102976091 | 0.32 |
ENST00000286918.4
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr19_+_17337027 | 0.32 |
ENST00000601529.1
ENST00000600232.1 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr3_-_107941209 | 0.32 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr7_-_6388389 | 0.31 |
ENST00000578372.1
|
FAM220A
|
family with sequence similarity 220, member A |
| chr3_-_119182453 | 0.31 |
ENST00000491685.1
ENST00000461654.1 |
TMEM39A
|
transmembrane protein 39A |
| chr12_-_92539614 | 0.31 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr7_-_86849836 | 0.31 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr17_-_4607335 | 0.30 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr6_-_31671058 | 0.30 |
ENST00000538874.1
ENST00000395952.3 |
ABHD16A
|
abhydrolase domain containing 16A |
| chr19_+_49122548 | 0.30 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr8_-_64080945 | 0.30 |
ENST00000603538.1
|
YTHDF3-AS1
|
YTHDF3 antisense RNA 1 (head to head) |
| chr14_+_102276192 | 0.30 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_26496362 | 0.29 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr11_+_65337901 | 0.29 |
ENST00000309328.3
ENST00000531405.1 ENST00000527920.1 ENST00000526877.1 ENST00000533115.1 ENST00000526433.1 |
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1 |
| chr8_+_98788057 | 0.29 |
ENST00000517924.1
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr6_-_27782548 | 0.28 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr1_+_6640046 | 0.28 |
ENST00000319084.5
ENST00000435905.1 |
ZBTB48
|
zinc finger and BTB domain containing 48 |
| chr7_-_150780609 | 0.28 |
ENST00000297533.4
|
TMUB1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr2_+_241375069 | 0.28 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr12_+_54378849 | 0.28 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr20_+_33292068 | 0.28 |
ENST00000374810.3
ENST00000374809.2 ENST00000451665.1 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr19_+_54704610 | 0.28 |
ENST00000302907.4
|
RPS9
|
ribosomal protein S9 |
| chr11_-_64889649 | 0.28 |
ENST00000434372.2
|
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr11_+_3819049 | 0.27 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr10_-_27444143 | 0.27 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr1_+_203274639 | 0.27 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr1_-_32110467 | 0.27 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr17_-_7155775 | 0.26 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr1_+_156698234 | 0.26 |
ENST00000368218.4
ENST00000368216.4 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chrX_-_77150911 | 0.26 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr1_+_156698708 | 0.26 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr2_-_98280383 | 0.26 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr19_+_55996565 | 0.26 |
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr17_-_43209862 | 0.26 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr7_+_139025875 | 0.26 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr5_-_162887054 | 0.25 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr17_-_77770830 | 0.25 |
ENST00000269385.4
|
CBX8
|
chromobox homolog 8 |
| chr16_-_67694597 | 0.25 |
ENST00000393919.4
ENST00000219251.8 |
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr17_-_43568062 | 0.25 |
ENST00000421073.2
ENST00000584420.1 ENST00000589780.1 ENST00000430334.3 |
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr5_+_139927213 | 0.25 |
ENST00000310331.2
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr7_-_45026419 | 0.25 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr19_+_36036477 | 0.24 |
ENST00000222284.5
ENST00000392204.2 |
TMEM147
|
transmembrane protein 147 |
| chr19_-_7553889 | 0.24 |
ENST00000221480.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr16_+_19078960 | 0.24 |
ENST00000568985.1
ENST00000566110.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr20_+_43343886 | 0.24 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr19_-_44100275 | 0.24 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr17_-_71308119 | 0.24 |
ENST00000439510.2
ENST00000581014.1 ENST00000579611.1 |
CDC42EP4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr17_+_73089382 | 0.24 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr16_+_2022036 | 0.24 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr8_-_29940569 | 0.24 |
ENST00000523761.1
|
TMEM66
|
transmembrane protein 66 |
| chr12_+_22778291 | 0.24 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr2_+_74781828 | 0.23 |
ENST00000340004.6
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr7_+_140396946 | 0.23 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr6_-_41888843 | 0.23 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr22_-_50683381 | 0.23 |
ENST00000439308.2
|
TUBGCP6
|
tubulin, gamma complex associated protein 6 |
| chr10_-_14920599 | 0.23 |
ENST00000609399.1
|
RP11-398C13.6
|
RP11-398C13.6 |
| chr5_+_174151536 | 0.23 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr19_+_54704718 | 0.23 |
ENST00000391752.1
ENST00000402367.1 ENST00000391751.3 |
RPS9
|
ribosomal protein S9 |
| chr1_-_153935791 | 0.23 |
ENST00000429040.1
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr12_+_51632666 | 0.23 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr3_-_134092561 | 0.22 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr7_-_45151272 | 0.22 |
ENST00000461363.1
ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chr9_-_34665983 | 0.22 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr16_+_67927147 | 0.22 |
ENST00000291041.5
|
PSKH1
|
protein serine kinase H1 |
| chr19_+_17337007 | 0.22 |
ENST00000215061.4
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr15_+_75287861 | 0.22 |
ENST00000425597.3
ENST00000562327.1 ENST00000568018.1 ENST00000562212.1 ENST00000567920.1 ENST00000566872.1 ENST00000361900.6 ENST00000545456.1 |
SCAMP5
|
secretory carrier membrane protein 5 |
| chr16_+_20912114 | 0.22 |
ENST00000567954.1
|
LYRM1
|
LYR motif containing 1 |
| chr4_-_140477910 | 0.22 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr7_+_100464760 | 0.22 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr19_+_36103631 | 0.22 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr3_-_49761337 | 0.21 |
ENST00000535833.1
ENST00000308388.6 ENST00000480687.1 ENST00000308375.6 |
AMIGO3
GMPPB
|
adhesion molecule with Ig-like domain 3 GDP-mannose pyrophosphorylase B |
| chr17_+_17380294 | 0.21 |
ENST00000268711.3
ENST00000580462.1 |
MED9
|
mediator complex subunit 9 |
| chr16_+_2039946 | 0.21 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr11_+_809647 | 0.21 |
ENST00000321153.4
|
RPLP2
|
ribosomal protein, large, P2 |
| chr4_+_119512928 | 0.21 |
ENST00000567913.2
|
RP11-384K6.6
|
RP11-384K6.6 |
| chr17_-_40828969 | 0.21 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr19_+_46009837 | 0.21 |
ENST00000589627.1
|
VASP
|
vasodilator-stimulated phosphoprotein |
| chr17_-_40829026 | 0.21 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr7_+_138915102 | 0.21 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr11_-_61584233 | 0.21 |
ENST00000491310.1
|
FADS1
|
fatty acid desaturase 1 |
| chr3_+_19988736 | 0.21 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr14_+_91526668 | 0.21 |
ENST00000521334.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr19_-_7553852 | 0.21 |
ENST00000593547.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr19_-_46088068 | 0.21 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr2_+_220143989 | 0.20 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr3_-_101395936 | 0.20 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr7_+_140396465 | 0.20 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr5_-_175964366 | 0.20 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr8_-_101964738 | 0.20 |
ENST00000523938.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr11_-_6633799 | 0.20 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chrX_-_106960285 | 0.20 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr19_+_54705025 | 0.20 |
ENST00000441429.1
|
RPS9
|
ribosomal protein S9 |
| chr19_-_55628927 | 0.20 |
ENST00000263433.3
ENST00000376393.2 |
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C |
| chr19_-_55919087 | 0.19 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr19_+_49122785 | 0.19 |
ENST00000598088.1
|
SPHK2
|
sphingosine kinase 2 |
| chr12_+_13044787 | 0.19 |
ENST00000534831.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chr6_-_32098013 | 0.19 |
ENST00000375156.3
|
FKBPL
|
FK506 binding protein like |
| chr5_+_133562095 | 0.19 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr19_+_50148087 | 0.19 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chr6_-_26285737 | 0.19 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr19_+_54704990 | 0.19 |
ENST00000391753.2
|
RPS9
|
ribosomal protein S9 |
| chr8_+_118533049 | 0.19 |
ENST00000522839.1
|
MED30
|
mediator complex subunit 30 |
| chr19_+_18723660 | 0.19 |
ENST00000262817.3
|
TMEM59L
|
transmembrane protein 59-like |
| chr11_-_114271139 | 0.19 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chrX_+_48398053 | 0.18 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr2_+_28974531 | 0.18 |
ENST00000420282.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr19_-_5720123 | 0.18 |
ENST00000587365.1
ENST00000585374.1 ENST00000593119.1 |
LONP1
|
lon peptidase 1, mitochondrial |
| chrX_-_153236819 | 0.18 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr10_+_99496872 | 0.18 |
ENST00000337540.7
ENST00000357540.4 ENST00000370613.3 ENST00000370610.3 ENST00000393677.4 ENST00000453958.2 ENST00000359980.3 |
ZFYVE27
|
zinc finger, FYVE domain containing 27 |
| chr11_+_394145 | 0.18 |
ENST00000528036.1
|
PKP3
|
plakophilin 3 |
| chr2_+_11052054 | 0.18 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr22_+_25003568 | 0.18 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr2_-_97535708 | 0.18 |
ENST00000305476.5
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr7_+_5632436 | 0.18 |
ENST00000340250.6
ENST00000382361.3 |
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chr3_-_156878482 | 0.18 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr6_-_38607628 | 0.18 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr19_+_44100632 | 0.18 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr19_+_18283959 | 0.18 |
ENST00000597802.2
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr22_+_25003606 | 0.17 |
ENST00000432867.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chrX_-_153237258 | 0.17 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr2_+_30454390 | 0.17 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr12_-_6982442 | 0.17 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr19_-_5720248 | 0.17 |
ENST00000360614.3
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr1_-_1284730 | 0.17 |
ENST00000378888.5
|
DVL1
|
dishevelled segment polarity protein 1 |
| chr16_+_88772866 | 0.17 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr3_-_48514604 | 0.17 |
ENST00000426002.1
|
SHISA5
|
shisa family member 5 |
| chr15_+_89182156 | 0.17 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr11_-_66056478 | 0.17 |
ENST00000431556.2
ENST00000528575.1 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr1_-_151254362 | 0.17 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr7_+_140396756 | 0.17 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr9_-_140082983 | 0.17 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chrX_+_152240819 | 0.17 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr11_+_12695944 | 0.17 |
ENST00000361905.4
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr16_-_30366672 | 0.17 |
ENST00000305596.3
|
CD2BP2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr10_-_53459319 | 0.16 |
ENST00000331173.4
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
| chr7_-_129592471 | 0.16 |
ENST00000473814.2
ENST00000490974.1 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr11_-_62389449 | 0.16 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr21_-_44846999 | 0.16 |
ENST00000270162.6
|
SIK1
|
salt-inducible kinase 1 |
| chr7_+_100273736 | 0.16 |
ENST00000412215.1
ENST00000393924.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr7_-_45026200 | 0.16 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr19_+_44100544 | 0.16 |
ENST00000391965.2
ENST00000525771.1 |
ZNF576
|
zinc finger protein 576 |
| chr4_-_54930790 | 0.16 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr2_+_220408724 | 0.16 |
ENST00000421791.1
ENST00000373883.3 ENST00000451952.1 |
TMEM198
|
transmembrane protein 198 |
| chr11_-_85376121 | 0.16 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr18_-_77711625 | 0.16 |
ENST00000357575.4
ENST00000590381.1 ENST00000397778.2 |
PQLC1
|
PQ loop repeat containing 1 |
| chr9_+_97488939 | 0.16 |
ENST00000277198.2
ENST00000297979.5 |
C9orf3
|
chromosome 9 open reading frame 3 |
| chr20_-_62258394 | 0.16 |
ENST00000370077.1
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr16_+_67261008 | 0.15 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr17_+_18163848 | 0.15 |
ENST00000323019.4
ENST00000578174.1 ENST00000395704.4 ENST00000395703.4 ENST00000578621.1 ENST00000579341.1 |
MIEF2
|
mitochondrial elongation factor 2 |
| chr12_+_98987369 | 0.15 |
ENST00000401722.3
ENST00000188376.5 ENST00000228318.3 ENST00000551917.1 ENST00000548046.1 ENST00000552981.1 ENST00000551265.1 ENST00000550695.1 ENST00000547534.1 ENST00000549338.1 ENST00000548847.1 |
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF2_ATF1_ATF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 0.5 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 0.9 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.6 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.7 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.4 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.5 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.4 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.4 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.3 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 1.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 0.8 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.1 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.6 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.3 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 1.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.2 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.0 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.4 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.2 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.5 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.7 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.0 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.2 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0008366 | ensheathment of neurons(GO:0007272) axon ensheathment(GO:0008366) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0098751 | megakaryocyte development(GO:0035855) bone cell development(GO:0098751) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.1 | GO:0052360 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0010868 | negative regulation of triglyceride biosynthetic process(GO:0010868) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0051664 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.0 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0052042 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.0 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) negative regulation of B cell differentiation(GO:0045578) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.0 | GO:0000806 | Y chromosome(GO:0000806) cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.0 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 0.5 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.2 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 0.5 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.5 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 0.4 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.7 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.3 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.2 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.2 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.2 | GO:0051908 | double-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0051908) |
| 0.0 | 0.1 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.4 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.0 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |