Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ARID5A
Z-value: 0.71
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.11 | AT-rich interaction domain 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg19_v2_chr2_+_97202480_97202499 | -0.89 | 1.1e-01 | Click! |
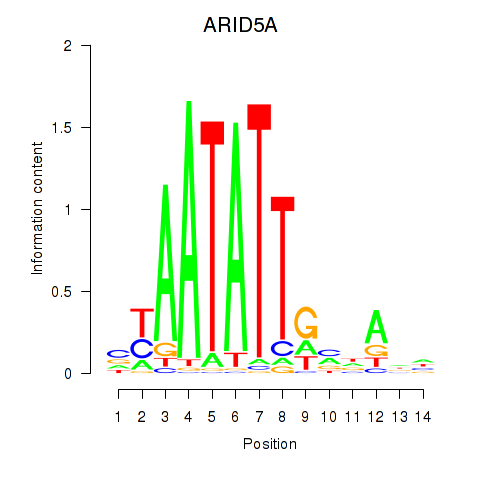
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_54267147 | 0.56 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr3_-_167191814 | 0.46 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chrX_-_55208866 | 0.46 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr12_-_11214893 | 0.46 |
ENST00000533467.1
|
TAS2R46
|
taste receptor, type 2, member 46 |
| chr6_+_26156551 | 0.41 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr6_-_26235206 | 0.37 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr14_+_56584414 | 0.34 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr9_-_3469181 | 0.32 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr5_+_159848854 | 0.29 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr18_+_68002675 | 0.27 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr1_+_104159999 | 0.27 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr2_+_90198535 | 0.26 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr3_-_196987309 | 0.26 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr6_+_26199737 | 0.26 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chrX_+_41583408 | 0.26 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr3_-_106959424 | 0.25 |
ENST00000607801.1
ENST00000479612.2 ENST00000484698.1 ENST00000477210.2 ENST00000473636.1 |
LINC00882
|
long intergenic non-protein coding RNA 882 |
| chr6_+_26104104 | 0.24 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr2_+_113321939 | 0.24 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr4_+_69962212 | 0.23 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr12_-_49333446 | 0.21 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr12_-_96389702 | 0.21 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr3_-_64253655 | 0.21 |
ENST00000498162.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr6_-_26056695 | 0.21 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr4_+_69962185 | 0.20 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr12_-_10588539 | 0.20 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr12_+_104337515 | 0.19 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr4_+_100737954 | 0.19 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr19_-_35264089 | 0.18 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chr3_-_150421728 | 0.18 |
ENST00000295910.6
ENST00000491361.1 |
FAM194A
|
family with sequence similarity 194, member A |
| chr22_-_32767017 | 0.18 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr19_-_44174330 | 0.18 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr22_-_32766972 | 0.18 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr21_-_35284635 | 0.17 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr6_+_76599809 | 0.17 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr19_-_48753028 | 0.17 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr8_-_128960591 | 0.17 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
| chr8_-_65730127 | 0.17 |
ENST00000522106.1
|
RP11-1D12.2
|
RP11-1D12.2 |
| chr4_-_90756769 | 0.16 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr19_-_44174305 | 0.16 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr1_-_113253977 | 0.16 |
ENST00000606505.1
ENST00000605933.1 |
RP11-426L16.10
|
Rho-related GTP-binding protein RhoC |
| chrX_+_70521584 | 0.16 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr5_+_159848807 | 0.16 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr8_-_112039643 | 0.16 |
ENST00000524283.1
|
RP11-946L20.2
|
RP11-946L20.2 |
| chrX_+_135252050 | 0.16 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr7_+_116593433 | 0.16 |
ENST00000323984.3
ENST00000393449.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_-_179457805 | 0.16 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr12_+_28605426 | 0.15 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr12_-_7596735 | 0.15 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr2_+_74120094 | 0.15 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr15_+_93749295 | 0.14 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chrX_-_100641155 | 0.14 |
ENST00000372880.1
ENST00000308731.7 |
BTK
|
Bruton agammaglobulinemia tyrosine kinase |
| chr16_-_67597789 | 0.14 |
ENST00000605277.1
|
CTD-2012K14.6
|
CTD-2012K14.6 |
| chr5_-_64064508 | 0.14 |
ENST00000513458.4
|
SREK1IP1
|
SREK1-interacting protein 1 |
| chr10_+_70847852 | 0.14 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr2_+_196313239 | 0.14 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr7_+_112262421 | 0.14 |
ENST00000453459.1
|
AC002463.3
|
AC002463.3 |
| chr2_-_188312971 | 0.13 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr5_+_61874562 | 0.13 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr11_+_128563652 | 0.13 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_+_31707725 | 0.13 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr11_+_55606228 | 0.13 |
ENST00000378396.1
|
OR5D16
|
olfactory receptor, family 5, subfamily D, member 16 |
| chr10_+_127661942 | 0.13 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr2_+_219646462 | 0.13 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr9_+_135937365 | 0.13 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr6_-_169654139 | 0.12 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chrX_+_23682379 | 0.12 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chr16_+_10479906 | 0.12 |
ENST00000562527.1
ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr3_-_69171708 | 0.12 |
ENST00000420581.2
|
LMOD3
|
leiomodin 3 (fetal) |
| chr16_+_14980632 | 0.12 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr2_+_109204909 | 0.12 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_-_125366018 | 0.12 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr7_+_116593292 | 0.12 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr22_+_25615489 | 0.12 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr18_-_50240 | 0.12 |
ENST00000573909.1
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr16_-_29910365 | 0.11 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr3_+_119217422 | 0.11 |
ENST00000466984.1
|
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr3_-_24207039 | 0.11 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr5_+_118690466 | 0.11 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr10_-_75118471 | 0.11 |
ENST00000340329.3
|
TTC18
|
tetratricopeptide repeat domain 18 |
| chr7_+_116593536 | 0.11 |
ENST00000417919.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr3_+_113616317 | 0.11 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr10_+_106113515 | 0.11 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr21_+_25801041 | 0.11 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr12_-_10542617 | 0.11 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr2_-_133429091 | 0.11 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr7_+_116654935 | 0.11 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_+_160336851 | 0.10 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr16_-_4588822 | 0.10 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr2_-_201753980 | 0.10 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr19_+_33865218 | 0.10 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chrX_+_22050546 | 0.10 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr11_+_57531292 | 0.10 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr11_-_4719072 | 0.10 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr1_+_116654376 | 0.10 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr6_-_74161977 | 0.10 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr12_+_4758264 | 0.10 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr2_+_219472488 | 0.10 |
ENST00000450993.2
|
PLCD4
|
phospholipase C, delta 4 |
| chr3_+_119217376 | 0.10 |
ENST00000494664.1
ENST00000493694.1 |
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chrX_+_107288239 | 0.09 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr7_+_107220422 | 0.09 |
ENST00000005259.4
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr6_-_127664683 | 0.09 |
ENST00000528402.1
ENST00000454591.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr10_+_90562705 | 0.09 |
ENST00000539337.1
|
LIPM
|
lipase, family member M |
| chr8_+_107738343 | 0.09 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr20_+_31870927 | 0.09 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr7_+_116593568 | 0.09 |
ENST00000446490.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr19_-_48752812 | 0.09 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr1_-_85358850 | 0.09 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr12_-_49582978 | 0.09 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr14_+_90722886 | 0.09 |
ENST00000543772.2
|
PSMC1
|
proteasome (prosome, macropain) 26S subunit, ATPase, 1 |
| chr12_+_70219052 | 0.09 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr2_-_214148921 | 0.09 |
ENST00000360083.3
|
AC079610.2
|
AC079610.2 |
| chr17_+_78518617 | 0.09 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr3_+_142342228 | 0.09 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr14_-_67878917 | 0.08 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr7_+_138943265 | 0.08 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr2_-_225362533 | 0.08 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr16_-_4588762 | 0.08 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr19_+_30156400 | 0.08 |
ENST00000588833.1
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr2_-_70520832 | 0.08 |
ENST00000454893.1
ENST00000272348.2 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr11_+_107879459 | 0.08 |
ENST00000393094.2
|
CUL5
|
cullin 5 |
| chr7_+_116502527 | 0.08 |
ENST00000361183.3
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr6_-_33239712 | 0.08 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr13_+_76445187 | 0.08 |
ENST00000318245.4
|
C13orf45
|
chromosome 13 open reading frame 45 |
| chr1_-_149459549 | 0.08 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr10_-_28623368 | 0.08 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr4_-_84035905 | 0.08 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr12_+_16500037 | 0.08 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chrX_-_45060135 | 0.08 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr21_+_17553910 | 0.08 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr10_-_61720640 | 0.08 |
ENST00000521074.1
ENST00000444900.1 |
C10orf40
|
chromosome 10 open reading frame 40 |
| chr12_+_21284118 | 0.08 |
ENST00000256958.2
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1 |
| chr11_-_112034803 | 0.07 |
ENST00000528832.1
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr22_+_32754139 | 0.07 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr8_+_87526732 | 0.07 |
ENST00000523469.1
ENST00000522240.1 |
CPNE3
|
copine III |
| chr7_-_91808441 | 0.07 |
ENST00000437357.1
ENST00000458448.1 |
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr7_+_98923505 | 0.07 |
ENST00000432884.2
ENST00000262942.5 |
ARPC1A
|
actin related protein 2/3 complex, subunit 1A, 41kDa |
| chr10_+_116697946 | 0.07 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr3_-_69129501 | 0.07 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr7_-_3214287 | 0.07 |
ENST00000404626.3
|
AC091801.1
|
LOC392621; Uncharacterized protein |
| chr12_+_131438443 | 0.07 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr3_+_184018352 | 0.07 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr1_+_247712383 | 0.07 |
ENST00000366488.4
ENST00000536561.1 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chrM_+_8366 | 0.07 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr6_-_138539627 | 0.07 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr11_-_73587719 | 0.07 |
ENST00000545127.1
ENST00000537289.1 ENST00000355693.4 |
COA4
|
cytochrome c oxidase assembly factor 4 homolog (S. cerevisiae) |
| chr5_-_76916396 | 0.07 |
ENST00000509971.1
|
WDR41
|
WD repeat domain 41 |
| chr11_+_73661364 | 0.07 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr7_-_103848405 | 0.07 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr18_+_52258390 | 0.07 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
| chr11_-_104480019 | 0.07 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr15_-_78913628 | 0.07 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chrX_+_107288197 | 0.06 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr19_+_36132631 | 0.06 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr11_+_76745385 | 0.06 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr15_-_55657428 | 0.06 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr8_-_124279627 | 0.06 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr11_+_60145948 | 0.06 |
ENST00000300184.3
ENST00000358246.1 |
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr4_-_69536346 | 0.06 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chrX_+_48755202 | 0.06 |
ENST00000447146.2
ENST00000376548.5 ENST00000247140.4 |
PQBP1
|
polyglutamine binding protein 1 |
| chr3_+_134204551 | 0.06 |
ENST00000332047.5
ENST00000354446.3 |
CEP63
|
centrosomal protein 63kDa |
| chr14_-_54425475 | 0.06 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr11_+_22688150 | 0.06 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr18_+_61575200 | 0.06 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr7_-_19748640 | 0.06 |
ENST00000222567.5
|
TWISTNB
|
TWIST neighbor |
| chr2_-_10587897 | 0.06 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr1_+_196743912 | 0.06 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr2_+_223725723 | 0.06 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr5_+_140165876 | 0.06 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr16_+_55542910 | 0.06 |
ENST00000262134.5
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2 |
| chr11_+_63655987 | 0.06 |
ENST00000509502.2
ENST00000512060.1 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr21_-_35899113 | 0.06 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr7_+_116502605 | 0.06 |
ENST00000458284.2
ENST00000490693.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr6_-_33239612 | 0.06 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr1_+_160175201 | 0.06 |
ENST00000368076.1
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr2_+_217277137 | 0.05 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr2_-_136594740 | 0.05 |
ENST00000264162.2
|
LCT
|
lactase |
| chr15_+_59063478 | 0.05 |
ENST00000559228.1
ENST00000450403.2 |
FAM63B
|
family with sequence similarity 63, member B |
| chr14_+_39703112 | 0.05 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr2_+_29353520 | 0.05 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr6_+_30035307 | 0.05 |
ENST00000376765.2
ENST00000376763.1 |
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr2_-_74007193 | 0.05 |
ENST00000377706.4
ENST00000443070.1 ENST00000272444.3 |
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr8_-_17942432 | 0.05 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr1_+_64014588 | 0.05 |
ENST00000371086.2
ENST00000340052.3 |
DLEU2L
|
deleted in lymphocytic leukemia 2-like |
| chrX_+_153991025 | 0.05 |
ENST00000369550.5
|
DKC1
|
dyskeratosis congenita 1, dyskerin |
| chr6_+_107077471 | 0.05 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr12_+_32260085 | 0.05 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr7_+_116654958 | 0.05 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr4_-_84035868 | 0.05 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr7_+_39125365 | 0.05 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr10_+_18689637 | 0.05 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr19_+_35417844 | 0.05 |
ENST00000601957.1
|
ZNF30
|
zinc finger protein 30 |
| chr9_-_140196703 | 0.05 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr1_+_212475148 | 0.05 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr11_-_62477041 | 0.05 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr19_+_35417798 | 0.05 |
ENST00000303586.7
ENST00000439785.1 ENST00000601540.1 |
ZNF30
|
zinc finger protein 30 |
| chr1_+_84609944 | 0.05 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr6_-_106773291 | 0.04 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr8_-_94029882 | 0.04 |
ENST00000520686.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr11_-_2170786 | 0.04 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_+_106959552 | 0.04 |
ENST00000473550.1
|
LINC00883
|
long intergenic non-protein coding RNA 883 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.3 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) B cell cytokine production(GO:0002368) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:1903284 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.3 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.3 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.0 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0072100 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.0 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.1 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.4 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.2 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.3 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |