Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ALX3
Z-value: 0.37
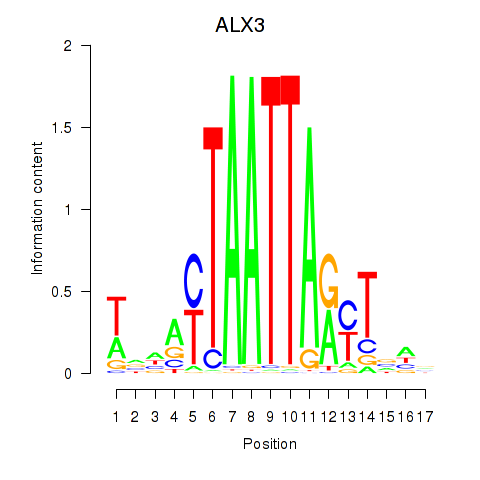
Transcription factors associated with ALX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX3
|
ENSG00000156150.6 | ALX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX3 | hg19_v2_chr1_-_110613276_110613322 | 0.28 | 7.2e-01 | Click! |
Activity profile of ALX3 motif
Sorted Z-values of ALX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_19091325 | 0.23 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr7_-_111032971 | 0.15 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr19_-_52307357 | 0.12 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr12_-_86650077 | 0.12 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr17_-_19015945 | 0.10 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr3_+_28390637 | 0.09 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr20_-_50419055 | 0.08 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr6_+_26183958 | 0.08 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr5_+_150639360 | 0.07 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr19_-_58864848 | 0.07 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr6_+_160542821 | 0.07 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr1_+_28261533 | 0.07 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr9_-_75488984 | 0.07 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr15_+_101417919 | 0.06 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr5_+_140227357 | 0.06 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr18_-_53303123 | 0.06 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr4_+_37455536 | 0.06 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr4_-_120243545 | 0.06 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr2_-_198540751 | 0.06 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr12_+_26164645 | 0.06 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr21_-_22175341 | 0.05 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr16_-_28634874 | 0.05 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_-_48116540 | 0.05 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr3_+_51851612 | 0.05 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr19_+_50016411 | 0.05 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr13_+_76413852 | 0.05 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr10_+_35894338 | 0.05 |
ENST00000321660.1
|
GJD4
|
gap junction protein, delta 4, 40.1kDa |
| chr4_-_89442940 | 0.05 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr11_+_5710919 | 0.04 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr4_+_144312659 | 0.04 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr12_-_10282836 | 0.04 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr4_-_103749205 | 0.04 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr19_+_50016610 | 0.04 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr7_+_133812052 | 0.04 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr20_-_50722183 | 0.04 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr1_+_225600404 | 0.04 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr20_-_50418972 | 0.04 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr12_+_78359999 | 0.04 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr16_-_28608364 | 0.04 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr17_-_7167279 | 0.04 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr17_+_35851570 | 0.04 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr2_+_13677795 | 0.04 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr4_+_169633310 | 0.04 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr7_-_22862406 | 0.04 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr17_+_12859080 | 0.03 |
ENST00000583608.1
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr11_+_100862811 | 0.03 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr8_-_90769422 | 0.03 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr11_-_62521614 | 0.03 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr20_-_43133491 | 0.03 |
ENST00000411544.1
|
SERINC3
|
serine incorporator 3 |
| chr12_-_25348007 | 0.03 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr1_+_84630574 | 0.03 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_+_110736659 | 0.03 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr15_-_37393406 | 0.03 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr11_-_107729287 | 0.03 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr9_+_111624577 | 0.03 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr17_-_4938712 | 0.03 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr1_+_47533160 | 0.03 |
ENST00000334194.3
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr2_+_102953608 | 0.03 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr7_-_111424506 | 0.03 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr1_+_151253991 | 0.03 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr6_+_26087646 | 0.03 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr12_-_46121554 | 0.03 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr2_-_152118352 | 0.03 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr16_-_28621353 | 0.03 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr8_-_623547 | 0.03 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr2_+_11682790 | 0.03 |
ENST00000389825.3
ENST00000381483.2 |
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr15_-_91565770 | 0.03 |
ENST00000535906.1
ENST00000333371.3 |
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr15_-_91565743 | 0.03 |
ENST00000535843.1
|
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr16_+_31271274 | 0.03 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr20_+_33134619 | 0.03 |
ENST00000374837.3
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr12_+_7014064 | 0.03 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr11_-_62323702 | 0.03 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr11_+_2405833 | 0.03 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr1_-_201140673 | 0.03 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr4_-_25865159 | 0.02 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr19_-_3557570 | 0.02 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr1_+_28261492 | 0.02 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr17_+_18086392 | 0.02 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr16_-_28937027 | 0.02 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr16_+_15489603 | 0.02 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr6_+_26087509 | 0.02 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr2_+_166095898 | 0.02 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr7_-_111424462 | 0.02 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr3_+_151591422 | 0.02 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr19_-_44388116 | 0.02 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr7_+_117864708 | 0.02 |
ENST00000357099.4
ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr16_-_28621312 | 0.02 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr11_+_114168085 | 0.02 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr8_-_124741451 | 0.02 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr5_+_176811431 | 0.02 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr6_+_26402465 | 0.02 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr7_+_37723420 | 0.02 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chr1_+_62439037 | 0.02 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr16_+_53133070 | 0.02 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_90112590 | 0.02 |
ENST00000472284.1
|
DAPK1
|
death-associated protein kinase 1 |
| chr11_-_107729504 | 0.02 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_-_103749179 | 0.02 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr9_+_90112117 | 0.02 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr4_+_86525299 | 0.02 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr9_+_34652164 | 0.02 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr17_-_19364269 | 0.02 |
ENST00000421796.2
ENST00000585389.1 ENST00000609249.1 |
AC004448.5
|
AC004448.5 |
| chr5_+_140201183 | 0.02 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr9_-_4666421 | 0.02 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr1_-_24126023 | 0.02 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr8_-_93978216 | 0.02 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr7_-_144435985 | 0.02 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr2_-_220264703 | 0.02 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr15_-_98417780 | 0.02 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr17_-_72772462 | 0.02 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_-_128894053 | 0.02 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr12_+_7014126 | 0.02 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr9_+_470288 | 0.02 |
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr1_+_157963063 | 0.02 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr7_-_87856303 | 0.02 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr3_+_186692745 | 0.02 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr15_-_75748143 | 0.02 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr1_-_17766198 | 0.02 |
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2 |
| chr15_+_58702742 | 0.02 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr4_-_103749105 | 0.02 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_-_39341594 | 0.02 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr21_+_33671160 | 0.02 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr13_+_24144796 | 0.02 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr12_+_64798826 | 0.02 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr19_-_36001113 | 0.02 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr7_-_87856280 | 0.02 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr7_-_43965937 | 0.02 |
ENST00000455877.1
ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP
|
upregulator of cell proliferation |
| chr19_+_48949030 | 0.02 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr19_+_49199209 | 0.02 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr1_-_92952433 | 0.02 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr9_+_12775011 | 0.02 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr5_+_140762268 | 0.02 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr16_-_28621298 | 0.01 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr12_-_10022735 | 0.01 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chrX_+_84258832 | 0.01 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr9_+_90112767 | 0.01 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr2_-_225811747 | 0.01 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr4_+_26344754 | 0.01 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_+_101918153 | 0.01 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr19_+_47421933 | 0.01 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr2_+_217524323 | 0.01 |
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr12_+_18414446 | 0.01 |
ENST00000433979.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr14_-_67878917 | 0.01 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr6_-_32095968 | 0.01 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr5_+_68860949 | 0.01 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr17_-_295730 | 0.01 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr5_+_66300446 | 0.01 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr22_+_17956618 | 0.01 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr9_+_90112741 | 0.01 |
ENST00000469640.2
|
DAPK1
|
death-associated protein kinase 1 |
| chr4_-_103749313 | 0.01 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_+_210517895 | 0.01 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr5_+_140579162 | 0.01 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr18_+_29171689 | 0.01 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr6_+_52442083 | 0.01 |
ENST00000606714.1
|
TRAM2-AS1
|
TRAM2 antisense RNA 1 (head to head) |
| chr5_-_94417186 | 0.01 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr17_-_72772425 | 0.01 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr3_-_126327398 | 0.01 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr3_+_139063372 | 0.01 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr2_-_74618964 | 0.01 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr10_-_99030395 | 0.01 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr10_+_6779326 | 0.01 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr10_+_5135981 | 0.01 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr16_-_66584059 | 0.01 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr3_+_119298523 | 0.01 |
ENST00000357003.3
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr11_+_100784231 | 0.01 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr1_+_144989309 | 0.01 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr5_-_20575959 | 0.01 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr2_-_228497888 | 0.01 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr11_-_559377 | 0.01 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr17_+_1674982 | 0.01 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr5_-_41794313 | 0.01 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr18_-_67624160 | 0.01 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr9_-_99540328 | 0.01 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr5_+_102200948 | 0.01 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr12_-_22063787 | 0.01 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr6_+_37897735 | 0.01 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr9_+_124329336 | 0.01 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr10_-_95241951 | 0.01 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr14_-_78083112 | 0.01 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr6_+_26440700 | 0.01 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr11_-_107729887 | 0.01 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr21_-_22175450 | 0.01 |
ENST00000435279.2
|
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr3_+_186353756 | 0.01 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr4_+_142142035 | 0.01 |
ENST00000262990.4
ENST00000512809.1 ENST00000503649.1 ENST00000512738.1 ENST00000421169.2 |
ZNF330
|
zinc finger protein 330 |
| chr10_-_95242044 | 0.01 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr2_-_74619152 | 0.01 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr14_+_32798462 | 0.01 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr6_+_3259148 | 0.01 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr20_-_56265680 | 0.01 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chrX_+_107288197 | 0.01 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr17_-_6524159 | 0.01 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr1_-_202897724 | 0.01 |
ENST00000435533.3
ENST00000367258.1 |
KLHL12
|
kelch-like family member 12 |
| chr2_-_101925055 | 0.01 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr10_-_75415825 | 0.01 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr17_+_40610862 | 0.01 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr9_-_5830768 | 0.01 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr15_+_78370140 | 0.01 |
ENST00000409568.2
|
SH2D7
|
SH2 domain containing 7 |
| chr8_+_29605841 | 0.01 |
ENST00000523123.1
|
AC145110.1
|
AC145110.1 |
| chrX_+_107288239 | 0.01 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_-_24126051 | 0.00 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr1_+_207226574 | 0.00 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.0 | GO:0046661 | sex differentiation(GO:0007548) gonad development(GO:0008406) male gonad development(GO:0008584) development of primary sexual characteristics(GO:0045137) development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |