Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
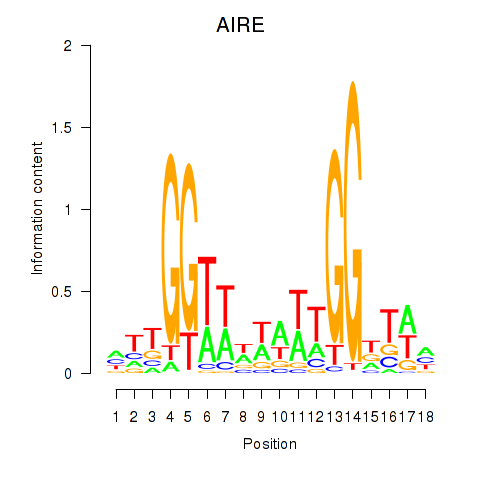
Results for AIRE
Z-value: 0.48
Transcription factors associated with AIRE
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AIRE
|
ENSG00000160224.12 | autoimmune regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AIRE | hg19_v2_chr21_+_45705752_45705767 | -0.03 | 9.7e-01 | Click! |
Activity profile of AIRE motif
Sorted Z-values of AIRE motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_56985873 | 0.28 |
ENST00000299721.3
|
CPLX4
|
complexin 4 |
| chrX_-_139866723 | 0.26 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr12_+_52643077 | 0.22 |
ENST00000553310.2
ENST00000544024.1 |
KRT86
|
keratin 86 |
| chr19_-_52552051 | 0.21 |
ENST00000221315.5
|
ZNF432
|
zinc finger protein 432 |
| chr6_-_33864684 | 0.21 |
ENST00000506222.2
ENST00000533304.1 |
LINC01016
|
long intergenic non-protein coding RNA 1016 |
| chr5_+_667759 | 0.20 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr14_-_75330537 | 0.17 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr12_+_75874580 | 0.16 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr19_-_52551814 | 0.16 |
ENST00000594154.1
ENST00000598745.1 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr2_+_38177575 | 0.16 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr22_-_32767017 | 0.16 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr7_-_64023441 | 0.15 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr19_-_23869970 | 0.14 |
ENST00000601010.1
|
ZNF675
|
zinc finger protein 675 |
| chr18_-_37380230 | 0.13 |
ENST00000591629.1
|
LINC00669
|
long intergenic non-protein coding RNA 669 |
| chr19_-_23869999 | 0.13 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr6_+_22221010 | 0.13 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr19_-_16606988 | 0.13 |
ENST00000269881.3
|
CALR3
|
calreticulin 3 |
| chr15_-_52043722 | 0.13 |
ENST00000454181.2
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr15_-_52043631 | 0.12 |
ENST00000558126.1
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr15_+_27112948 | 0.11 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr21_+_30502806 | 0.11 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr1_+_165864821 | 0.11 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr16_-_20566616 | 0.10 |
ENST00000569163.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr16_+_75182376 | 0.10 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr4_-_88450612 | 0.10 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr10_+_90660832 | 0.10 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr22_+_42017987 | 0.09 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr6_-_31514333 | 0.09 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr10_+_115511213 | 0.09 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr19_-_48753028 | 0.08 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr9_+_42717234 | 0.08 |
ENST00000377590.1
|
FOXD4L2
|
forkhead box D4-like 2 |
| chr2_+_62423242 | 0.08 |
ENST00000301998.4
|
B3GNT2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chr4_-_4544061 | 0.07 |
ENST00000507908.1
|
STX18
|
syntaxin 18 |
| chr5_+_122181279 | 0.07 |
ENST00000395451.4
ENST00000506996.1 |
SNX24
|
sorting nexin 24 |
| chr5_+_122181184 | 0.07 |
ENST00000513881.1
|
SNX24
|
sorting nexin 24 |
| chr9_-_114361665 | 0.07 |
ENST00000309195.5
|
PTGR1
|
prostaglandin reductase 1 |
| chr4_+_71587669 | 0.07 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr2_+_172864490 | 0.07 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr12_+_20963647 | 0.07 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr4_-_104119528 | 0.07 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr9_-_70429731 | 0.07 |
ENST00000377413.1
|
FOXD4L4
|
forkhead box D4-like 4 |
| chr15_-_38519066 | 0.07 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr9_-_70178815 | 0.06 |
ENST00000377420.1
|
FOXD4L5
|
forkhead box D4-like 5 |
| chr15_+_85523671 | 0.06 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr3_+_140981456 | 0.06 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr14_+_39944025 | 0.06 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr10_+_127661942 | 0.06 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr10_-_69597810 | 0.06 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr2_-_127977654 | 0.06 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr12_+_4758264 | 0.06 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr8_+_124780672 | 0.06 |
ENST00000521166.1
ENST00000334705.7 |
FAM91A1
|
family with sequence similarity 91, member A1 |
| chr2_-_198540719 | 0.05 |
ENST00000295049.4
|
RFTN2
|
raftlin family member 2 |
| chr1_+_174670143 | 0.05 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr9_-_104198042 | 0.05 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr8_+_128748308 | 0.05 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr8_+_104384616 | 0.05 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr6_+_168434678 | 0.05 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr9_-_95298254 | 0.05 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr5_+_118690466 | 0.05 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr15_+_52043758 | 0.05 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chrX_-_102941596 | 0.05 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr13_-_26795840 | 0.05 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr16_-_31147020 | 0.05 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr6_-_106773610 | 0.05 |
ENST00000369076.3
ENST00000369070.1 |
ATG5
|
autophagy related 5 |
| chr10_-_101841588 | 0.05 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr12_+_10658489 | 0.05 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr10_+_115511434 | 0.05 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr2_+_179316163 | 0.05 |
ENST00000409117.3
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr12_+_75874984 | 0.05 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr3_-_121468602 | 0.05 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chrM_+_12331 | 0.05 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr1_-_145826450 | 0.04 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr12_-_76462713 | 0.04 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr4_+_71588372 | 0.04 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr7_-_64467031 | 0.04 |
ENST00000394323.2
|
ERV3-1
|
endogenous retrovirus group 3, member 1 |
| chr1_+_36038971 | 0.04 |
ENST00000373235.3
|
TFAP2E
|
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
| chr4_+_76649797 | 0.04 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr14_-_55658252 | 0.04 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chrM_+_5824 | 0.04 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr11_+_64794991 | 0.04 |
ENST00000352068.5
ENST00000525648.1 |
SNX15
|
sorting nexin 15 |
| chr20_+_21284416 | 0.04 |
ENST00000539513.1
|
XRN2
|
5'-3' exoribonuclease 2 |
| chr7_-_137028498 | 0.04 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr7_-_137028534 | 0.04 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr5_+_122181128 | 0.04 |
ENST00000261369.4
|
SNX24
|
sorting nexin 24 |
| chr18_-_56985776 | 0.04 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr14_+_45605157 | 0.04 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr6_+_26104104 | 0.04 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr6_+_118869452 | 0.04 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr4_-_47465666 | 0.04 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr17_+_77030267 | 0.04 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr1_-_52344471 | 0.04 |
ENST00000352171.7
ENST00000354831.7 |
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr12_+_75874460 | 0.03 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr3_-_133648656 | 0.03 |
ENST00000408895.2
|
C3orf36
|
chromosome 3 open reading frame 36 |
| chr9_-_25678856 | 0.03 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr19_-_48752812 | 0.03 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr20_+_44036900 | 0.03 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr14_-_55658323 | 0.03 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr9_-_114362093 | 0.03 |
ENST00000538962.1
ENST00000407693.2 ENST00000238248.3 |
PTGR1
|
prostaglandin reductase 1 |
| chr17_+_3323862 | 0.03 |
ENST00000291231.1
|
OR3A3
|
olfactory receptor, family 3, subfamily A, member 3 |
| chr3_-_121468513 | 0.03 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr1_-_52344416 | 0.03 |
ENST00000544028.1
|
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr8_-_141774467 | 0.03 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr2_-_136633940 | 0.03 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chrX_-_139047669 | 0.03 |
ENST00000370540.1
|
CXorf66
|
chromosome X open reading frame 66 |
| chr19_-_48753104 | 0.03 |
ENST00000447740.2
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr4_+_78829479 | 0.03 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr22_+_25202232 | 0.03 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr6_+_26538566 | 0.03 |
ENST00000377575.2
|
HMGN4
|
high mobility group nucleosomal binding domain 4 |
| chr1_+_47603109 | 0.03 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr7_-_87936195 | 0.03 |
ENST00000414498.1
ENST00000301959.5 ENST00000380079.4 |
STEAP4
|
STEAP family member 4 |
| chr14_-_65409438 | 0.03 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr17_+_41857793 | 0.03 |
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105 |
| chr2_-_166810261 | 0.03 |
ENST00000243344.7
|
TTC21B
|
tetratricopeptide repeat domain 21B |
| chr16_-_89119355 | 0.03 |
ENST00000537498.1
|
CTD-2555A7.2
|
CTD-2555A7.2 |
| chr22_+_40322623 | 0.03 |
ENST00000399090.2
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr22_+_40322595 | 0.03 |
ENST00000420971.1
ENST00000544756.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr1_-_154600421 | 0.02 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr5_+_141488070 | 0.02 |
ENST00000253814.4
|
NDFIP1
|
Nedd4 family interacting protein 1 |
| chr13_+_50570019 | 0.02 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr12_-_64784471 | 0.02 |
ENST00000333722.5
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr6_+_42584847 | 0.02 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr2_-_225811747 | 0.02 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr14_+_45605127 | 0.02 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr1_-_35497506 | 0.02 |
ENST00000317538.5
ENST00000373340.2 ENST00000357182.4 |
ZMYM6
|
zinc finger, MYM-type 6 |
| chr6_+_116937636 | 0.02 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr16_+_50059125 | 0.02 |
ENST00000427478.2
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr3_-_121264848 | 0.02 |
ENST00000264233.5
|
POLQ
|
polymerase (DNA directed), theta |
| chr2_-_202508169 | 0.02 |
ENST00000409883.2
|
TMEM237
|
transmembrane protein 237 |
| chr4_-_114682719 | 0.02 |
ENST00000394522.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chrM_+_14741 | 0.02 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr11_-_119249805 | 0.02 |
ENST00000527843.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr12_-_59175485 | 0.02 |
ENST00000550678.1
ENST00000552201.1 |
RP11-767I20.1
|
RP11-767I20.1 |
| chr1_+_207669573 | 0.02 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr1_+_207669613 | 0.02 |
ENST00000367049.4
ENST00000529814.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr19_+_45417921 | 0.02 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr2_+_204732487 | 0.02 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr4_+_1385340 | 0.02 |
ENST00000324803.4
|
CRIPAK
|
cysteine-rich PAK1 inhibitor |
| chr4_-_66536057 | 0.02 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr19_+_11708259 | 0.01 |
ENST00000587939.1
ENST00000588174.1 |
ZNF627
|
zinc finger protein 627 |
| chr16_+_14802801 | 0.01 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr6_+_167525277 | 0.01 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chrM_+_8366 | 0.01 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr12_-_50677255 | 0.01 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr1_+_100810575 | 0.01 |
ENST00000542213.1
|
CDC14A
|
cell division cycle 14A |
| chr19_-_36304201 | 0.01 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr10_-_4720333 | 0.01 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr9_-_113761720 | 0.01 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr13_+_111893533 | 0.01 |
ENST00000478679.1
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr6_-_149806105 | 0.01 |
ENST00000389942.5
ENST00000416573.2 ENST00000542614.1 ENST00000409806.3 |
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr21_+_39644395 | 0.01 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr8_-_141728444 | 0.01 |
ENST00000521562.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr1_-_167883327 | 0.01 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr21_+_39644214 | 0.01 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_-_52456352 | 0.01 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr9_+_71986182 | 0.01 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr3_+_189507460 | 0.01 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr21_+_39644172 | 0.00 |
ENST00000398932.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr20_+_44036620 | 0.00 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr4_-_66536196 | 0.00 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr2_+_234686976 | 0.00 |
ENST00000389758.3
|
MROH2A
|
maestro heat-like repeat family member 2A |
| chr17_+_36452989 | 0.00 |
ENST00000312513.5
ENST00000582535.1 |
MRPL45
|
mitochondrial ribosomal protein L45 |
| chr5_+_72143988 | 0.00 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr6_+_42141029 | 0.00 |
ENST00000372958.1
|
GUCA1A
|
guanylate cyclase activator 1A (retina) |
| chr4_-_17513851 | 0.00 |
ENST00000281243.5
|
QDPR
|
quinoid dihydropteridine reductase |
| chr21_+_39644305 | 0.00 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AIRE
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.1 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.0 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) all-trans retinal binding(GO:0005503) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.0 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |