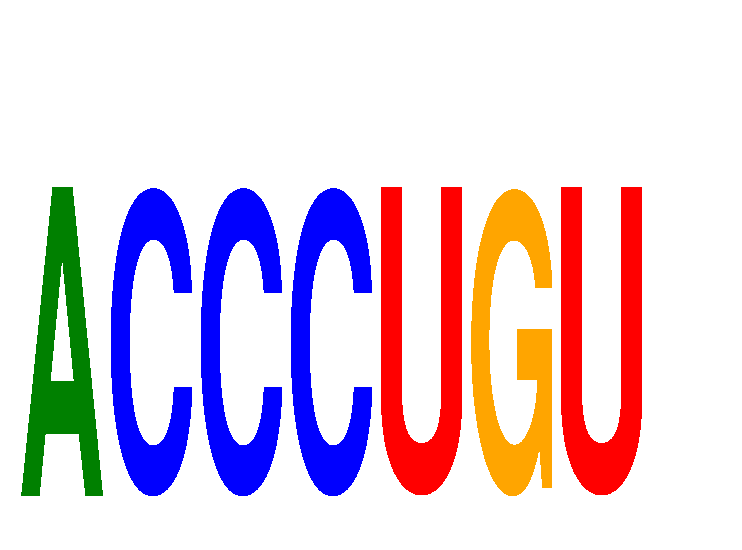Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ACCCUGU
Z-value: 0.07
miRNA associated with seed ACCCUGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-10a-5p
|
MIMAT0000253 |
|
hsa-miR-10b-5p
|
MIMAT0000254 |
Activity profile of ACCCUGU motif
Sorted Z-values of ACCCUGU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_176981307 | 0.02 |
ENST00000249501.4
|
HOXD10
|
homeobox D10 |
| chr2_+_10183651 | 0.02 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr14_+_45366472 | 0.01 |
ENST00000325192.3
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr3_-_72496035 | 0.01 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chr2_+_64681219 | 0.01 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr11_+_18344106 | 0.01 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr6_-_146135880 | 0.01 |
ENST00000237281.4
|
FBXO30
|
F-box protein 30 |
| chrX_+_117629766 | 0.01 |
ENST00000276204.6
ENST00000276202.7 |
DOCK11
|
dedicator of cytokinesis 11 |
| chr7_+_94139105 | 0.01 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr1_-_85156216 | 0.01 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr17_-_29151794 | 0.01 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr16_+_69599861 | 0.01 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr1_+_210406121 | 0.01 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr3_+_178866199 | 0.01 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr5_-_90679145 | 0.01 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr3_-_142166904 | 0.01 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr2_+_238536207 | 0.01 |
ENST00000308482.9
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chr18_+_43914159 | 0.01 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr10_+_72972281 | 0.01 |
ENST00000335350.6
|
UNC5B
|
unc-5 homolog B (C. elegans) |
| chr17_+_45727204 | 0.01 |
ENST00000290158.4
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr10_+_180987 | 0.01 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr7_-_72936531 | 0.01 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr4_-_77135046 | 0.01 |
ENST00000264896.2
|
SCARB2
|
scavenger receptor class B, member 2 |
| chr1_-_39339777 | 0.01 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr5_+_167181917 | 0.01 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr5_+_63461642 | 0.01 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr1_+_28052456 | 0.01 |
ENST00000373954.6
ENST00000419687.2 |
FAM76A
|
family with sequence similarity 76, member A |
| chr22_+_45559722 | 0.01 |
ENST00000347635.4
ENST00000407019.2 ENST00000424634.1 ENST00000417702.1 ENST00000425733.2 ENST00000430547.1 |
NUP50
|
nucleoporin 50kDa |
| chr6_-_11044509 | 0.01 |
ENST00000354666.3
|
ELOVL2
|
ELOVL fatty acid elongase 2 |
| chr1_+_57110972 | 0.01 |
ENST00000371244.4
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr18_+_19749386 | 0.01 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr10_-_128994422 | 0.01 |
ENST00000522781.1
|
FAM196A
|
family with sequence similarity 196, member A |
| chr18_-_45456930 | 0.01 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr11_-_18656028 | 0.01 |
ENST00000336349.5
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
| chr5_+_149109825 | 0.01 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr19_-_55954230 | 0.01 |
ENST00000376325.4
|
SHISA7
|
shisa family member 7 |
| chr13_-_29069232 | 0.00 |
ENST00000282397.4
ENST00000541932.1 ENST00000539099.1 |
FLT1
|
fms-related tyrosine kinase 1 |
| chr12_-_29534074 | 0.00 |
ENST00000546839.1
ENST00000360150.4 ENST00000552155.1 ENST00000550353.1 ENST00000548441.1 ENST00000552132.1 |
ERGIC2
|
ERGIC and golgi 2 |
| chr1_+_109632425 | 0.00 |
ENST00000338272.8
|
TMEM167B
|
transmembrane protein 167B |
| chr10_-_88281494 | 0.00 |
ENST00000298767.5
|
WAPAL
|
wings apart-like homolog (Drosophila) |
| chr9_+_129567282 | 0.00 |
ENST00000449886.1
ENST00000373464.4 ENST00000450858.1 |
ZBTB43
|
zinc finger and BTB domain containing 43 |
| chr1_-_94374946 | 0.00 |
ENST00000370238.3
|
GCLM
|
glutamate-cysteine ligase, modifier subunit |
| chr15_-_56209306 | 0.00 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr5_+_179921430 | 0.00 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr1_+_93544791 | 0.00 |
ENST00000545708.1
ENST00000540243.1 ENST00000370298.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr10_+_8096631 | 0.00 |
ENST00000379328.3
|
GATA3
|
GATA binding protein 3 |
| chr1_-_19283163 | 0.00 |
ENST00000455833.2
|
IFFO2
|
intermediate filament family orphan 2 |
| chr2_+_23608064 | 0.00 |
ENST00000486442.1
|
KLHL29
|
kelch-like family member 29 |
| chr2_-_206950781 | 0.00 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr13_+_98086445 | 0.00 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr8_-_41655107 | 0.00 |
ENST00000347528.4
ENST00000289734.7 ENST00000379758.2 ENST00000396945.1 ENST00000396942.1 ENST00000352337.4 |
ANK1
|
ankyrin 1, erythrocytic |
| chr22_+_39898325 | 0.00 |
ENST00000325301.2
ENST00000404569.1 |
MIEF1
|
mitochondrial elongation factor 1 |
| chr4_-_66536057 | 0.00 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr17_-_49198216 | 0.00 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr5_+_56205081 | 0.00 |
ENST00000285947.2
ENST00000541720.1 |
SETD9
|
SET domain containing 9 |
| chr20_+_55204351 | 0.00 |
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr6_-_10415470 | 0.00 |
ENST00000379604.2
ENST00000379613.3 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr12_+_4918342 | 0.00 |
ENST00000280684.3
ENST00000433855.1 |
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6 |
| chr21_-_32931290 | 0.00 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr14_-_48143999 | 0.00 |
ENST00000439988.3
|
MDGA2
|
MAM domain-containing glycosylphosphatidylinositol anchor protein 2 |
| chr8_-_4852218 | 0.00 |
ENST00000400186.3
ENST00000602723.1 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr18_-_33647487 | 0.00 |
ENST00000590898.1
ENST00000357384.4 ENST00000319040.6 ENST00000588737.1 ENST00000399022.4 |
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr20_+_31407692 | 0.00 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr10_-_30348439 | 0.00 |
ENST00000375377.1
|
KIAA1462
|
KIAA1462 |
| chr16_+_50187556 | 0.00 |
ENST00000561678.1
ENST00000357464.3 |
PAPD5
|
PAP associated domain containing 5 |
| chr10_-_121632266 | 0.00 |
ENST00000360003.3
ENST00000369077.3 |
MCMBP
|
minichromosome maintenance complex binding protein |
| chr7_-_138666053 | 0.00 |
ENST00000440172.1
ENST00000422774.1 |
KIAA1549
|
KIAA1549 |
| chr1_+_110082487 | 0.00 |
ENST00000527748.1
|
GPR61
|
G protein-coupled receptor 61 |
| chr19_+_10527449 | 0.00 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr1_+_202976493 | 0.00 |
ENST00000367242.3
|
TMEM183A
|
transmembrane protein 183A |
| chr6_+_71377473 | 0.00 |
ENST00000370452.3
ENST00000316999.5 ENST00000370455.3 |
SMAP1
|
small ArfGAP 1 |
| chr10_-_75173785 | 0.00 |
ENST00000535178.1
ENST00000372921.5 ENST00000372919.4 |
ANXA7
|
annexin A7 |
| chr4_-_152147579 | 0.00 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr4_-_111119804 | 0.00 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr1_-_38325256 | 0.00 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr12_-_77459306 | 0.00 |
ENST00000547316.1
ENST00000416496.2 ENST00000550669.1 ENST00000322886.7 |
E2F7
|
E2F transcription factor 7 |
| chrX_-_54384425 | 0.00 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr6_+_116421976 | 0.00 |
ENST00000319550.4
ENST00000419791.1 |
NT5DC1
|
5'-nucleotidase domain containing 1 |
| chr11_+_65101225 | 0.00 |
ENST00000528416.1
ENST00000415073.2 ENST00000252268.4 |
DPF2
|
D4, zinc and double PHD fingers family 2 |
| chr7_-_112430647 | 0.00 |
ENST00000312814.6
|
TMEM168
|
transmembrane protein 168 |
| chr9_+_100263912 | 0.00 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr11_+_66025167 | 0.00 |
ENST00000394067.2
ENST00000316924.5 ENST00000421552.1 ENST00000394078.1 |
KLC2
|
kinesin light chain 2 |
| chr1_+_169337172 | 0.00 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr4_+_47033345 | 0.00 |
ENST00000295454.3
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr6_+_117002339 | 0.00 |
ENST00000413340.1
ENST00000368564.1 ENST00000356348.1 |
KPNA5
|
karyopherin alpha 5 (importin alpha 6) |
| chr1_+_89990431 | 0.00 |
ENST00000330947.2
ENST00000358200.4 |
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr20_-_5591626 | 0.00 |
ENST00000379019.4
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr16_-_70719925 | 0.00 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr12_-_14956396 | 0.00 |
ENST00000535328.1
ENST00000261167.2 |
WBP11
|
WW domain binding protein 11 |
| chr1_+_224301787 | 0.00 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chr10_+_103113802 | 0.00 |
ENST00000370187.3
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr1_+_145477060 | 0.00 |
ENST00000369308.3
|
LIX1L
|
Lix1 homolog (mouse)-like |
| chr9_-_95527079 | 0.00 |
ENST00000356884.6
ENST00000375512.3 |
BICD2
|
bicaudal D homolog 2 (Drosophila) |
| chr22_-_17602200 | 0.00 |
ENST00000399875.1
|
CECR6
|
cat eye syndrome chromosome region, candidate 6 |
| chr17_+_34058639 | 0.00 |
ENST00000268864.3
|
RASL10B
|
RAS-like, family 10, member B |
| chr10_+_98592009 | 0.00 |
ENST00000540664.1
ENST00000371103.3 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr8_-_30891078 | 0.00 |
ENST00000339382.2
ENST00000475541.1 |
PURG
|
purine-rich element binding protein G |
| chr2_-_69870835 | 0.00 |
ENST00000409085.4
ENST00000406297.3 |
AAK1
|
AP2 associated kinase 1 |
| chr17_-_56084578 | 0.00 |
ENST00000582730.2
ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1
|
serine/arginine-rich splicing factor 1 |
| chr16_-_85722530 | 0.00 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr17_+_44928946 | 0.00 |
ENST00000290015.2
ENST00000393461.2 |
WNT9B
|
wingless-type MMTV integration site family, member 9B |
| chr7_-_44365020 | 0.00 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr2_+_148778570 | 0.00 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr7_-_92463210 | 0.00 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chr17_-_73775839 | 0.00 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr3_-_179754706 | 0.00 |
ENST00000465751.1
ENST00000467460.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr2_+_208394616 | 0.00 |
ENST00000432329.2
ENST00000353267.3 ENST00000445803.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr14_+_33408449 | 0.00 |
ENST00000346562.2
ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr19_+_3880581 | 0.00 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr4_+_79697495 | 0.00 |
ENST00000502871.1
ENST00000335016.5 |
BMP2K
|
BMP2 inducible kinase |
| chrX_-_46618490 | 0.00 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr17_-_79479789 | 0.00 |
ENST00000571691.1
ENST00000571721.1 ENST00000573283.1 ENST00000575842.1 ENST00000575087.1 ENST00000570382.1 ENST00000331925.2 |
ACTG1
|
actin, gamma 1 |