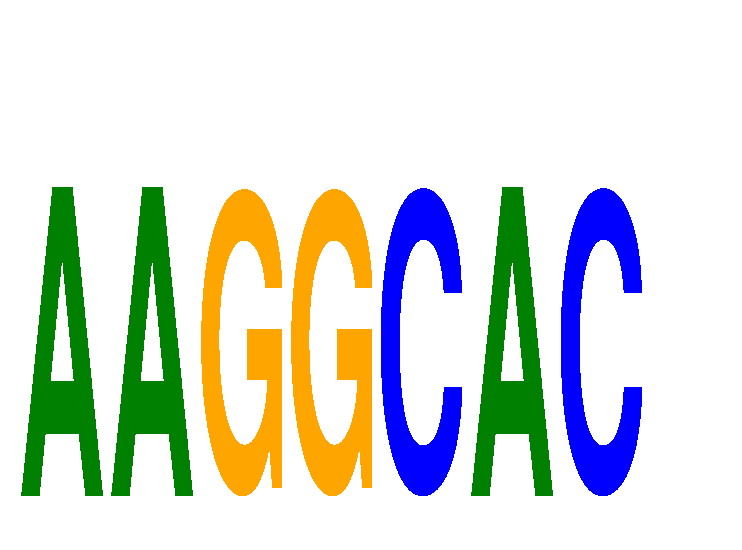Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for AAGGCAC
Z-value: 0.65
miRNA associated with seed AAGGCAC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-124-3p.1
|
MIMAT0000422 |
Activity profile of AAGGCAC motif
Sorted Z-values of AAGGCAC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_67236691 | 0.18 |
ENST00000544903.1
ENST00000308022.2 ENST00000393877.3 ENST00000452789.2 |
TMEM134
|
transmembrane protein 134 |
| chr3_-_32022733 | 0.17 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chr1_-_179834311 | 0.17 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr17_-_7297833 | 0.16 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr22_+_50354104 | 0.16 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr11_+_46402583 | 0.16 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr8_+_143808605 | 0.15 |
ENST00000336138.3
|
THEM6
|
thioesterase superfamily member 6 |
| chr22_+_21996549 | 0.14 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr11_-_45687128 | 0.14 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr17_-_17494972 | 0.14 |
ENST00000435340.2
ENST00000255389.5 ENST00000395781.2 |
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr5_+_172068232 | 0.14 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr1_-_72748417 | 0.14 |
ENST00000357731.5
|
NEGR1
|
neuronal growth regulator 1 |
| chr20_-_32031680 | 0.14 |
ENST00000217381.2
|
SNTA1
|
syntrophin, alpha 1 |
| chr17_-_41174424 | 0.13 |
ENST00000355653.3
|
VAT1
|
vesicle amine transport 1 |
| chr22_-_50946113 | 0.13 |
ENST00000216080.5
ENST00000474879.2 ENST00000380796.3 |
LMF2
|
lipase maturation factor 2 |
| chr9_-_111929560 | 0.13 |
ENST00000561981.2
|
FRRS1L
|
ferric-chelate reductase 1-like |
| chr6_+_100054606 | 0.13 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr2_-_73340146 | 0.13 |
ENST00000258098.6
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I) |
| chr11_-_3862206 | 0.13 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr12_+_52445191 | 0.12 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr19_-_16738984 | 0.12 |
ENST00000600060.1
ENST00000263390.3 |
MED26
|
mediator complex subunit 26 |
| chr2_-_85829811 | 0.12 |
ENST00000306353.3
|
TMEM150A
|
transmembrane protein 150A |
| chr1_+_31885963 | 0.12 |
ENST00000373709.3
|
SERINC2
|
serine incorporator 2 |
| chr1_+_179712298 | 0.12 |
ENST00000341785.4
|
FAM163A
|
family with sequence similarity 163, member A |
| chr1_+_26856236 | 0.12 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr2_+_241508039 | 0.12 |
ENST00000270357.4
|
RNPEPL1
|
arginyl aminopeptidase (aminopeptidase B)-like 1 |
| chr22_-_26986045 | 0.12 |
ENST00000442495.1
ENST00000440953.1 ENST00000450022.1 ENST00000338754.4 |
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr19_+_13842559 | 0.12 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr17_-_61777459 | 0.11 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr17_-_42402138 | 0.11 |
ENST00000592857.1
ENST00000586016.1 ENST00000590194.1 ENST00000377095.5 ENST00000588049.1 ENST00000586633.1 ENST00000537904.2 ENST00000585636.1 ENST00000585523.1 ENST00000225308.8 |
SLC25A39
|
solute carrier family 25, member 39 |
| chr19_-_11450249 | 0.11 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr11_+_64126614 | 0.11 |
ENST00000528057.1
ENST00000334205.4 ENST00000294261.4 |
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr16_-_122619 | 0.11 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr17_+_30593195 | 0.11 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr9_+_71394945 | 0.11 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr1_-_16482554 | 0.11 |
ENST00000358432.5
|
EPHA2
|
EPH receptor A2 |
| chr16_+_3184924 | 0.11 |
ENST00000574902.1
ENST00000396878.3 |
ZNF213
|
zinc finger protein 213 |
| chr9_+_123970052 | 0.11 |
ENST00000373823.3
|
GSN
|
gelsolin |
| chr9_+_131549483 | 0.11 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr9_+_140500087 | 0.11 |
ENST00000371421.4
|
ARRDC1
|
arrestin domain containing 1 |
| chr3_+_69812877 | 0.11 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr11_+_120081475 | 0.11 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr18_-_53255766 | 0.11 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr7_-_27183263 | 0.11 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr6_-_169654139 | 0.11 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr14_-_21566731 | 0.10 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr14_-_77495007 | 0.10 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr8_+_35649365 | 0.10 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr6_-_41909561 | 0.10 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr3_-_48936272 | 0.10 |
ENST00000544097.1
ENST00000430379.1 ENST00000319017.4 |
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr9_-_110251836 | 0.10 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr12_-_53625958 | 0.10 |
ENST00000327550.3
ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG
|
retinoic acid receptor, gamma |
| chr11_+_76494253 | 0.10 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr2_+_241375069 | 0.10 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr22_-_50746027 | 0.10 |
ENST00000425954.1
ENST00000449103.1 |
PLXNB2
|
plexin B2 |
| chr1_+_895930 | 0.10 |
ENST00000338591.3
|
KLHL17
|
kelch-like family member 17 |
| chr6_-_28220002 | 0.10 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chrX_+_153686614 | 0.09 |
ENST00000369682.3
|
PLXNA3
|
plexin A3 |
| chr10_-_50323543 | 0.09 |
ENST00000332853.4
ENST00000298454.3 |
VSTM4
|
V-set and transmembrane domain containing 4 |
| chr1_-_204121013 | 0.09 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_-_179846928 | 0.09 |
ENST00000367612.3
ENST00000609928.1 |
TOR1AIP2
|
torsin A interacting protein 2 |
| chr11_+_832944 | 0.09 |
ENST00000322008.4
ENST00000397421.1 ENST00000529810.1 ENST00000526693.1 ENST00000525333.1 ENST00000524748.1 ENST00000527341.1 |
CD151
|
CD151 molecule (Raph blood group) |
| chr11_+_818902 | 0.09 |
ENST00000336615.4
|
PNPLA2
|
patatin-like phospholipase domain containing 2 |
| chr12_-_124018252 | 0.09 |
ENST00000376874.4
|
RILPL1
|
Rab interacting lysosomal protein-like 1 |
| chr1_-_234745234 | 0.09 |
ENST00000366610.3
ENST00000366609.3 |
IRF2BP2
|
interferon regulatory factor 2 binding protein 2 |
| chr17_+_73512594 | 0.09 |
ENST00000333213.6
|
TSEN54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr19_-_45908292 | 0.08 |
ENST00000360957.5
ENST00000592134.1 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chrX_+_72783026 | 0.08 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr1_-_118472216 | 0.08 |
ENST00000369443.5
|
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr10_-_106098162 | 0.08 |
ENST00000337478.1
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr16_+_28303804 | 0.08 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr19_-_1863567 | 0.08 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr17_-_42908155 | 0.08 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr14_+_23067146 | 0.08 |
ENST00000428304.2
|
ABHD4
|
abhydrolase domain containing 4 |
| chr15_+_79724858 | 0.08 |
ENST00000305428.3
|
KIAA1024
|
KIAA1024 |
| chr1_-_1535455 | 0.08 |
ENST00000422725.1
|
C1orf233
|
chromosome 1 open reading frame 233 |
| chr20_+_4129426 | 0.08 |
ENST00000339123.6
ENST00000305958.4 ENST00000278795.3 |
SMOX
|
spermine oxidase |
| chr2_+_65216462 | 0.08 |
ENST00000234256.3
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr8_-_141645645 | 0.08 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr17_-_202579 | 0.08 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr3_-_13461807 | 0.08 |
ENST00000254508.5
|
NUP210
|
nucleoporin 210kDa |
| chr16_-_2246436 | 0.08 |
ENST00000343516.6
|
CASKIN1
|
CASK interacting protein 1 |
| chr7_-_100860851 | 0.08 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr14_-_99737565 | 0.08 |
ENST00000357195.3
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr9_-_135996537 | 0.08 |
ENST00000372050.3
ENST00000372047.3 |
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr4_+_7045042 | 0.08 |
ENST00000310074.7
ENST00000512388.1 |
TADA2B
|
transcriptional adaptor 2B |
| chr9_-_91793675 | 0.08 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr19_-_3063099 | 0.08 |
ENST00000221561.8
|
AES
|
amino-terminal enhancer of split |
| chr1_-_249120832 | 0.08 |
ENST00000366472.5
|
SH3BP5L
|
SH3-binding domain protein 5-like |
| chr10_-_15413035 | 0.08 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chr2_+_73441350 | 0.08 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr8_+_37654424 | 0.08 |
ENST00000315215.7
|
GPR124
|
G protein-coupled receptor 124 |
| chr12_+_48166978 | 0.07 |
ENST00000442218.2
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr18_+_905104 | 0.07 |
ENST00000579794.1
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary) |
| chr1_+_15736359 | 0.07 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr8_-_134584152 | 0.07 |
ENST00000521180.1
ENST00000517668.1 ENST00000319914.5 |
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr22_+_40573921 | 0.07 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr14_-_74551172 | 0.07 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr1_+_36348790 | 0.07 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr5_+_137801160 | 0.07 |
ENST00000239938.4
|
EGR1
|
early growth response 1 |
| chr10_+_98741041 | 0.07 |
ENST00000286067.2
|
C10orf12
|
chromosome 10 open reading frame 12 |
| chr19_+_2785458 | 0.07 |
ENST00000307741.6
ENST00000585338.1 |
THOP1
|
thimet oligopeptidase 1 |
| chr4_+_108910870 | 0.07 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr2_+_27070964 | 0.07 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr1_+_180123969 | 0.07 |
ENST00000367602.3
ENST00000367600.5 |
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr19_+_676385 | 0.07 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr16_-_84651673 | 0.07 |
ENST00000262428.4
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chr20_-_14318248 | 0.07 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr12_-_76478686 | 0.07 |
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr14_+_21538429 | 0.07 |
ENST00000298694.4
ENST00000555038.1 |
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr1_+_955448 | 0.07 |
ENST00000379370.2
|
AGRN
|
agrin |
| chr11_+_73019282 | 0.07 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr2_+_28615669 | 0.07 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr1_-_55352834 | 0.07 |
ENST00000371269.3
|
DHCR24
|
24-dehydrocholesterol reductase |
| chr1_+_179050512 | 0.07 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chr11_-_60929074 | 0.07 |
ENST00000301765.5
|
VPS37C
|
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
| chr5_+_137774706 | 0.07 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr17_+_72772621 | 0.07 |
ENST00000335464.5
ENST00000417024.2 ENST00000578764.1 ENST00000582773.1 ENST00000582330.1 |
TMEM104
|
transmembrane protein 104 |
| chr6_-_107436473 | 0.07 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr19_-_14117074 | 0.07 |
ENST00000588885.1
ENST00000254325.4 |
RFX1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr22_+_50624323 | 0.07 |
ENST00000380909.4
ENST00000303434.4 |
TRABD
|
TraB domain containing |
| chr6_-_134639180 | 0.07 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr18_+_19749386 | 0.07 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr10_-_64576105 | 0.07 |
ENST00000242480.3
ENST00000411732.1 |
EGR2
|
early growth response 2 |
| chr11_+_68816356 | 0.07 |
ENST00000294309.3
ENST00000542467.1 |
TPCN2
|
two pore segment channel 2 |
| chr19_+_3880581 | 0.07 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr15_-_55562582 | 0.07 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_+_52404270 | 0.07 |
ENST00000552049.1
ENST00000546756.1 |
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
| chr2_+_26395939 | 0.07 |
ENST00000401533.2
|
GAREML
|
GRB2 associated, regulator of MAPK1-like |
| chr10_+_18549645 | 0.07 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr6_-_33756867 | 0.07 |
ENST00000293760.5
|
LEMD2
|
LEM domain containing 2 |
| chr6_+_157802165 | 0.07 |
ENST00000414563.2
ENST00000359775.5 |
ZDHHC14
|
zinc finger, DHHC-type containing 14 |
| chr15_+_40763150 | 0.07 |
ENST00000306243.5
ENST00000559991.1 |
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr2_-_97405775 | 0.06 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr1_-_207224307 | 0.06 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr6_+_143929307 | 0.06 |
ENST00000427704.2
ENST00000305766.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr20_-_10654639 | 0.06 |
ENST00000254958.5
|
JAG1
|
jagged 1 |
| chr1_+_214161272 | 0.06 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr17_-_41856305 | 0.06 |
ENST00000397937.2
ENST00000226004.3 |
DUSP3
|
dual specificity phosphatase 3 |
| chr9_-_130341268 | 0.06 |
ENST00000373314.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chr10_-_126432619 | 0.06 |
ENST00000337318.3
|
FAM53B
|
family with sequence similarity 53, member B |
| chr10_-_99531709 | 0.06 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr20_+_62496596 | 0.06 |
ENST00000369927.4
ENST00000346249.4 ENST00000348257.5 ENST00000352482.4 ENST00000351424.4 ENST00000217121.5 ENST00000358548.4 |
TPD52L2
|
tumor protein D52-like 2 |
| chr1_+_11751748 | 0.06 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr11_+_60681346 | 0.06 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr7_+_138916231 | 0.06 |
ENST00000473989.3
ENST00000288561.8 |
UBN2
|
ubinuclein 2 |
| chr12_-_65146636 | 0.06 |
ENST00000418919.2
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr22_+_29279552 | 0.06 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr9_-_101984184 | 0.06 |
ENST00000476832.1
|
ALG2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr3_+_51575596 | 0.06 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr22_+_18121562 | 0.06 |
ENST00000355028.3
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr1_+_160370344 | 0.06 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr11_-_63439013 | 0.06 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr1_+_160336851 | 0.06 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr5_-_96478466 | 0.06 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr1_-_200992827 | 0.06 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr19_+_58838369 | 0.06 |
ENST00000329665.4
|
ZSCAN22
|
zinc finger and SCAN domain containing 22 |
| chr10_+_63661053 | 0.06 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr11_+_33278811 | 0.06 |
ENST00000303296.4
ENST00000379016.3 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr2_-_71221942 | 0.06 |
ENST00000272438.4
|
TEX261
|
testis expressed 261 |
| chr1_-_101360331 | 0.06 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr18_+_43914159 | 0.06 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr7_+_98972298 | 0.06 |
ENST00000252725.5
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa |
| chr15_-_51058005 | 0.06 |
ENST00000261854.5
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr2_-_230933709 | 0.06 |
ENST00000436869.1
ENST00000295190.4 |
SLC16A14
|
solute carrier family 16, member 14 |
| chr16_+_69221028 | 0.06 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chrX_-_77150985 | 0.06 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chrX_+_153607557 | 0.06 |
ENST00000369842.4
ENST00000369835.3 |
EMD
|
emerin |
| chr19_+_54371114 | 0.06 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr11_+_63580855 | 0.06 |
ENST00000294244.4
|
C11orf84
|
chromosome 11 open reading frame 84 |
| chr1_+_220701456 | 0.06 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr1_+_28052456 | 0.06 |
ENST00000373954.6
ENST00000419687.2 |
FAM76A
|
family with sequence similarity 76, member A |
| chr7_-_106301405 | 0.06 |
ENST00000523505.1
|
CCDC71L
|
coiled-coil domain containing 71-like |
| chr11_+_65154070 | 0.06 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr1_-_21671968 | 0.06 |
ENST00000415912.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr12_-_58146048 | 0.06 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr9_-_123639600 | 0.06 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr5_-_101632153 | 0.06 |
ENST00000310954.6
|
SLCO4C1
|
solute carrier organic anion transporter family, member 4C1 |
| chr5_+_175792459 | 0.06 |
ENST00000310389.5
|
ARL10
|
ADP-ribosylation factor-like 10 |
| chr6_+_138725343 | 0.05 |
ENST00000607197.1
ENST00000367697.3 |
HEBP2
|
heme binding protein 2 |
| chrX_-_134186144 | 0.05 |
ENST00000370775.2
|
FAM127B
|
family with sequence similarity 127, member B |
| chr15_-_75871589 | 0.05 |
ENST00000306726.2
|
PTPN9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr22_+_31090793 | 0.05 |
ENST00000332585.6
ENST00000382310.3 ENST00000446658.2 |
OSBP2
|
oxysterol binding protein 2 |
| chr3_-_48700310 | 0.05 |
ENST00000164024.4
ENST00000544264.1 |
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr7_-_124405681 | 0.05 |
ENST00000303921.2
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr1_+_25870070 | 0.05 |
ENST00000374338.4
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1 |
| chr20_-_30795511 | 0.05 |
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2 |
| chr1_+_160175117 | 0.05 |
ENST00000360472.4
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr8_-_20161466 | 0.05 |
ENST00000381569.1
|
LZTS1
|
leucine zipper, putative tumor suppressor 1 |
| chr17_+_2240775 | 0.05 |
ENST00000268989.3
ENST00000426855.2 |
SGSM2
|
small G protein signaling modulator 2 |
| chr19_-_36523709 | 0.05 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr7_-_112430647 | 0.05 |
ENST00000312814.6
|
TMEM168
|
transmembrane protein 168 |
| chr9_+_112403088 | 0.05 |
ENST00000448454.2
|
PALM2
|
paralemmin 2 |
| chr9_+_126773880 | 0.05 |
ENST00000373615.4
|
LHX2
|
LIM homeobox 2 |
| chr12_+_119616447 | 0.05 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr5_+_154092396 | 0.05 |
ENST00000336314.4
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr21_-_44846999 | 0.05 |
ENST00000270162.6
|
SIK1
|
salt-inducible kinase 1 |
| chr5_+_131705438 | 0.05 |
ENST00000245407.3
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr3_-_126076264 | 0.05 |
ENST00000296233.3
|
KLF15
|
Kruppel-like factor 15 |
| chrX_-_54384425 | 0.05 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGCAC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.1 | GO:0045362 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.0 | 0.1 | GO:0035284 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0002877 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.0 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.0 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.0 | GO:0060922 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.0 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.0 | GO:1905044 | Schwann cell proliferation involved in axon regeneration(GO:0014011) negative regulation of Schwann cell migration(GO:1900148) regulation of Schwann cell proliferation involved in axon regeneration(GO:1905044) negative regulation of Schwann cell proliferation involved in axon regeneration(GO:1905045) |
| 0.0 | 0.0 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.0 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) sequestering of neurotransmitter(GO:0042137) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.0 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.0 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.0 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.0 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.0 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |