Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for AACAGUC
Z-value: 0.35
miRNA associated with seed AACAGUC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-132-3p
|
MIMAT0000426 |
|
hsa-miR-212-3p
|
MIMAT0000269 |
Activity profile of AACAGUC motif
Sorted Z-values of AACAGUC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_34931458 | 0.11 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chrY_+_14813160 | 0.08 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr5_+_157170703 | 0.07 |
ENST00000286307.5
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chr3_+_197687071 | 0.07 |
ENST00000482695.1
ENST00000330198.4 ENST00000419117.1 ENST00000420910.2 ENST00000332636.5 |
LMLN
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr5_-_98262240 | 0.07 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr5_+_138678131 | 0.07 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr11_-_32452357 | 0.06 |
ENST00000379079.2
ENST00000530998.1 |
WT1
|
Wilms tumor 1 |
| chr5_+_137801160 | 0.06 |
ENST00000239938.4
|
EGR1
|
early growth response 1 |
| chr12_-_76953284 | 0.06 |
ENST00000547544.1
ENST00000393249.2 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr2_+_157291953 | 0.06 |
ENST00000310454.6
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr1_-_91487013 | 0.06 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr12_+_4918342 | 0.05 |
ENST00000280684.3
ENST00000433855.1 |
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6 |
| chr18_-_45456930 | 0.05 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr1_+_97187318 | 0.05 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr5_+_86564739 | 0.05 |
ENST00000456692.2
ENST00000512763.1 ENST00000506290.1 |
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr5_+_60628074 | 0.05 |
ENST00000252744.5
|
ZSWIM6
|
zinc finger, SWIM-type containing 6 |
| chr6_-_111136513 | 0.05 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr5_-_114880533 | 0.04 |
ENST00000274457.3
|
FEM1C
|
fem-1 homolog c (C. elegans) |
| chr17_+_21279509 | 0.04 |
ENST00000583088.1
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chrX_-_135056216 | 0.04 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr2_+_48010221 | 0.04 |
ENST00000234420.5
|
MSH6
|
mutS homolog 6 |
| chr18_-_29264669 | 0.04 |
ENST00000306851.5
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr15_+_44719394 | 0.04 |
ENST00000260327.4
ENST00000396780.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr10_+_5454505 | 0.04 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr11_+_34073195 | 0.04 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr18_-_23670546 | 0.04 |
ENST00000542743.1
ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr6_-_132272504 | 0.04 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr3_+_180630090 | 0.04 |
ENST00000357559.4
ENST00000305586.7 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr17_+_57784826 | 0.04 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr14_-_30396948 | 0.04 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr11_+_18344106 | 0.04 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr15_-_69113218 | 0.04 |
ENST00000560303.1
ENST00000465139.2 |
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr2_-_39348137 | 0.04 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr7_+_12250886 | 0.04 |
ENST00000444443.1
ENST00000396667.3 |
TMEM106B
|
transmembrane protein 106B |
| chr14_-_58894332 | 0.04 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_-_59225844 | 0.04 |
ENST00000380516.2
|
SLTM
|
SAFB-like, transcription modulator |
| chr15_+_38544476 | 0.03 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr4_+_111397216 | 0.03 |
ENST00000265162.5
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A) |
| chr17_-_16118835 | 0.03 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr3_-_9291063 | 0.03 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr11_+_119019722 | 0.03 |
ENST00000307417.3
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr13_+_48877895 | 0.03 |
ENST00000267163.4
|
RB1
|
retinoblastoma 1 |
| chr2_+_32288725 | 0.03 |
ENST00000315285.3
|
SPAST
|
spastin |
| chr9_+_15553055 | 0.03 |
ENST00000380701.3
|
CCDC171
|
coiled-coil domain containing 171 |
| chr1_-_23670817 | 0.03 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr22_+_35653445 | 0.03 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr1_-_53018654 | 0.03 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr13_-_41240717 | 0.03 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr8_-_119634141 | 0.03 |
ENST00000409003.4
ENST00000526328.1 ENST00000314727.4 ENST00000526765.1 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr4_+_52709229 | 0.03 |
ENST00000334635.5
ENST00000381441.3 ENST00000381437.4 |
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chr5_+_61602055 | 0.03 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr9_-_88714421 | 0.03 |
ENST00000388712.3
|
GOLM1
|
golgi membrane protein 1 |
| chr8_-_11324273 | 0.03 |
ENST00000284486.4
|
FAM167A
|
family with sequence similarity 167, member A |
| chr15_+_49715293 | 0.03 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr18_-_18691739 | 0.03 |
ENST00000399799.2
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr13_-_50510434 | 0.03 |
ENST00000361840.3
|
SPRYD7
|
SPRY domain containing 7 |
| chr9_+_103204553 | 0.03 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr3_+_62304712 | 0.03 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr4_+_48343339 | 0.03 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr5_-_139726181 | 0.03 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr17_-_65362678 | 0.03 |
ENST00000357146.4
ENST00000356126.3 |
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr10_+_88516396 | 0.03 |
ENST00000372037.3
|
BMPR1A
|
bone morphogenetic protein receptor, type IA |
| chr1_+_39456895 | 0.03 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr1_+_222791417 | 0.03 |
ENST00000344922.5
ENST00000344441.6 ENST00000344507.1 |
MIA3
|
melanoma inhibitory activity family, member 3 |
| chr18_+_9913977 | 0.03 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr17_-_40833858 | 0.03 |
ENST00000332438.4
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr16_-_47007545 | 0.03 |
ENST00000317089.5
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr1_-_235491462 | 0.03 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr2_+_85198216 | 0.03 |
ENST00000456682.1
ENST00000409785.4 |
KCMF1
|
potassium channel modulatory factor 1 |
| chr7_-_26904317 | 0.03 |
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr6_+_42749759 | 0.02 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr7_+_114562172 | 0.02 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr20_-_60640866 | 0.02 |
ENST00000252996.4
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr1_-_57889687 | 0.02 |
ENST00000371236.2
ENST00000371230.1 |
DAB1
|
Dab, reelin signal transducer, homolog 1 (Drosophila) |
| chr22_+_41487711 | 0.02 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr10_-_15210666 | 0.02 |
ENST00000378165.4
|
NMT2
|
N-myristoyltransferase 2 |
| chr14_-_90085458 | 0.02 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr2_+_85360499 | 0.02 |
ENST00000282111.3
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr8_-_74884511 | 0.02 |
ENST00000518127.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr16_-_46865047 | 0.02 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr9_+_103235365 | 0.02 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr10_-_32636106 | 0.02 |
ENST00000263062.8
ENST00000319778.6 |
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr7_-_75988321 | 0.02 |
ENST00000307630.3
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
| chr14_-_31676964 | 0.02 |
ENST00000553700.1
|
HECTD1
|
HECT domain containing E3 ubiquitin protein ligase 1 |
| chr1_-_244013384 | 0.02 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr7_-_111846435 | 0.02 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr2_-_65357225 | 0.02 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr13_+_114238997 | 0.02 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr9_+_129622904 | 0.02 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34 |
| chr6_+_110501344 | 0.02 |
ENST00000368932.1
|
CDC40
|
cell division cycle 40 |
| chr2_-_151344172 | 0.02 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr12_+_66217911 | 0.02 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr4_+_144257915 | 0.02 |
ENST00000262995.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr5_+_14664762 | 0.02 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr10_-_102027420 | 0.02 |
ENST00000354105.4
|
CWF19L1
|
CWF19-like 1, cell cycle control (S. pombe) |
| chr5_+_153825510 | 0.02 |
ENST00000297109.6
|
SAP30L
|
SAP30-like |
| chr1_-_169863016 | 0.02 |
ENST00000367772.4
ENST00000367771.6 |
SCYL3
|
SCY1-like 3 (S. cerevisiae) |
| chr8_-_30670384 | 0.02 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr10_+_114709999 | 0.02 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr6_+_111195973 | 0.02 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr3_+_178866199 | 0.02 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr6_+_87865262 | 0.02 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr12_+_68042495 | 0.02 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr1_+_57110972 | 0.02 |
ENST00000371244.4
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr7_+_94139105 | 0.02 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr15_+_50716576 | 0.02 |
ENST00000560297.1
ENST00000307179.4 ENST00000396444.3 ENST00000433963.1 ENST00000425032.3 |
USP8
|
ubiquitin specific peptidase 8 |
| chr13_-_77900814 | 0.02 |
ENST00000544440.2
|
MYCBP2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr8_-_41909496 | 0.02 |
ENST00000265713.2
ENST00000406337.1 ENST00000396930.3 ENST00000485568.1 ENST00000426524.1 |
KAT6A
|
K(lysine) acetyltransferase 6A |
| chr16_+_83841448 | 0.02 |
ENST00000433866.2
|
HSBP1
|
heat shock factor binding protein 1 |
| chr3_+_169684553 | 0.02 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chrX_+_12156582 | 0.02 |
ENST00000380682.1
|
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr14_+_102228123 | 0.01 |
ENST00000422945.2
ENST00000554442.1 ENST00000556260.2 ENST00000328724.5 ENST00000557268.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr4_+_144106080 | 0.01 |
ENST00000307017.4
|
USP38
|
ubiquitin specific peptidase 38 |
| chr11_-_12030629 | 0.01 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr1_-_93645818 | 0.01 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr3_+_37493610 | 0.01 |
ENST00000264741.5
|
ITGA9
|
integrin, alpha 9 |
| chr7_+_128379346 | 0.01 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr17_-_48785216 | 0.01 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr2_-_157189180 | 0.01 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr10_+_93683519 | 0.01 |
ENST00000265990.6
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chr4_+_26862400 | 0.01 |
ENST00000467011.1
ENST00000412829.2 |
STIM2
|
stromal interaction molecule 2 |
| chr12_+_69633317 | 0.01 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr12_+_62654119 | 0.01 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr22_-_37099555 | 0.01 |
ENST00000300105.6
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr8_+_133787586 | 0.01 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr5_-_132299313 | 0.01 |
ENST00000265343.5
|
AFF4
|
AF4/FMR2 family, member 4 |
| chr10_-_131762105 | 0.01 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr11_-_47198380 | 0.01 |
ENST00000419701.2
ENST00000526342.1 ENST00000528444.1 ENST00000530596.1 ENST00000525398.1 ENST00000319543.6 ENST00000426335.2 ENST00000527927.1 ENST00000525314.1 |
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr17_-_73775839 | 0.01 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr14_+_39644387 | 0.01 |
ENST00000553331.1
ENST00000216832.4 |
PNN
|
pinin, desmosome associated protein |
| chr6_+_163835669 | 0.01 |
ENST00000453779.2
ENST00000275262.7 ENST00000392127.2 ENST00000361752.3 |
QKI
|
QKI, KH domain containing, RNA binding |
| chr6_+_13615554 | 0.01 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chr4_+_124320665 | 0.01 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr3_+_107241783 | 0.01 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr4_-_1714037 | 0.01 |
ENST00000488267.1
ENST00000429429.2 ENST00000480936.1 |
SLBP
|
stem-loop binding protein |
| chr17_-_41277467 | 0.01 |
ENST00000494123.1
ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1
|
breast cancer 1, early onset |
| chr7_-_139876812 | 0.01 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr12_+_70636765 | 0.01 |
ENST00000552231.1
ENST00000229195.3 ENST00000547780.1 ENST00000418359.3 |
CNOT2
|
CCR4-NOT transcription complex, subunit 2 |
| chr18_-_5895954 | 0.01 |
ENST00000581347.2
|
TMEM200C
|
transmembrane protein 200C |
| chr14_-_64010046 | 0.01 |
ENST00000337537.3
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr10_+_114206956 | 0.01 |
ENST00000432306.1
ENST00000393077.2 |
VTI1A
|
vesicle transport through interaction with t-SNAREs 1A |
| chr15_-_31283798 | 0.01 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr4_-_120550146 | 0.01 |
ENST00000354960.3
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr1_+_209859510 | 0.01 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr1_-_154842741 | 0.01 |
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr7_+_107384142 | 0.01 |
ENST00000440859.3
|
CBLL1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr3_+_139654018 | 0.01 |
ENST00000458420.3
|
CLSTN2
|
calsyntenin 2 |
| chr5_-_131826457 | 0.01 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr4_+_57138437 | 0.01 |
ENST00000504228.1
ENST00000541073.1 |
KIAA1211
|
KIAA1211 |
| chr8_+_67579807 | 0.01 |
ENST00000519289.1
ENST00000519561.1 ENST00000521889.1 |
C8orf44-SGK3
C8orf44
|
C8orf44-SGK3 readthrough chromosome 8 open reading frame 44 |
| chr12_-_72057638 | 0.01 |
ENST00000552037.1
ENST00000378743.3 |
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr9_-_16870704 | 0.01 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr2_+_32390925 | 0.01 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr5_+_78532003 | 0.01 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr9_+_115249100 | 0.01 |
ENST00000337530.6
ENST00000374244.3 ENST00000536272.1 |
KIAA1958
|
KIAA1958 |
| chr9_+_128024067 | 0.01 |
ENST00000461379.1
ENST00000394084.1 ENST00000394105.2 ENST00000470056.1 ENST00000394104.2 ENST00000265956.4 ENST00000394083.2 ENST00000495955.1 ENST00000467750.1 ENST00000297933.6 |
GAPVD1
|
GTPase activating protein and VPS9 domains 1 |
| chr5_-_111093406 | 0.01 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein |
| chr20_+_32077880 | 0.01 |
ENST00000342704.6
ENST00000375279.2 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chrX_+_40944871 | 0.01 |
ENST00000378308.2
ENST00000324545.8 |
USP9X
|
ubiquitin specific peptidase 9, X-linked |
| chr11_-_70507901 | 0.01 |
ENST00000449833.2
ENST00000357171.3 ENST00000449116.2 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr22_+_29168652 | 0.01 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr8_+_38614807 | 0.01 |
ENST00000330691.6
ENST00000348567.4 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr14_-_61190754 | 0.01 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr1_+_200708671 | 0.01 |
ENST00000358823.2
|
CAMSAP2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr9_-_3525968 | 0.01 |
ENST00000382004.3
ENST00000302303.1 ENST00000449190.1 |
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr3_-_119813264 | 0.01 |
ENST00000264235.8
|
GSK3B
|
glycogen synthase kinase 3 beta |
| chr6_+_84569359 | 0.01 |
ENST00000369681.5
ENST00000369679.4 |
CYB5R4
|
cytochrome b5 reductase 4 |
| chr14_-_21905395 | 0.01 |
ENST00000430710.3
ENST00000553283.1 |
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chr12_+_113229737 | 0.01 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr15_+_79575073 | 0.01 |
ENST00000421388.2
|
ANKRD34C
|
ankyrin repeat domain 34C |
| chr10_-_13390270 | 0.01 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr17_+_30677136 | 0.01 |
ENST00000394670.4
ENST00000321233.6 ENST00000394673.2 ENST00000341711.6 ENST00000579634.1 ENST00000580759.1 ENST00000342555.6 ENST00000577908.1 ENST00000394679.5 ENST00000582165.1 |
ZNF207
|
zinc finger protein 207 |
| chr4_+_699537 | 0.01 |
ENST00000419774.1
ENST00000362003.5 ENST00000400151.2 ENST00000427463.1 ENST00000470161.2 |
PCGF3
|
polycomb group ring finger 3 |
| chr5_-_83680603 | 0.01 |
ENST00000296591.5
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr20_+_57226284 | 0.01 |
ENST00000458280.1
ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16
|
syntaxin 16 |
| chr5_+_122424816 | 0.01 |
ENST00000407847.4
|
PRDM6
|
PR domain containing 6 |
| chr14_-_35182994 | 0.01 |
ENST00000341223.3
|
CFL2
|
cofilin 2 (muscle) |
| chr1_+_215256467 | 0.01 |
ENST00000391894.2
ENST00000444842.2 |
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr6_-_139695757 | 0.01 |
ENST00000367651.2
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr3_+_186501336 | 0.01 |
ENST00000323963.5
ENST00000440191.2 ENST00000356531.5 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr3_-_122233723 | 0.01 |
ENST00000493510.1
ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1
|
karyopherin alpha 1 (importin alpha 5) |
| chr1_-_115300592 | 0.01 |
ENST00000261443.5
ENST00000534699.1 ENST00000339438.6 ENST00000529046.1 ENST00000525970.1 ENST00000369530.1 ENST00000530886.1 |
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr14_-_38064198 | 0.01 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr14_-_95786200 | 0.01 |
ENST00000298912.4
|
CLMN
|
calmin (calponin-like, transmembrane) |
| chr14_-_93799360 | 0.01 |
ENST00000334746.5
ENST00000554565.1 ENST00000298896.3 |
BTBD7
|
BTB (POZ) domain containing 7 |
| chr13_+_100258907 | 0.00 |
ENST00000376355.3
ENST00000376360.1 ENST00000444838.2 ENST00000376354.1 ENST00000339105.4 |
CLYBL
|
citrate lyase beta like |
| chr10_+_69644404 | 0.00 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr1_+_244816237 | 0.00 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr15_+_41709302 | 0.00 |
ENST00000389629.4
|
RTF1
|
Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr5_-_150948414 | 0.00 |
ENST00000261800.5
|
FAT2
|
FAT atypical cadherin 2 |
| chr16_+_69599861 | 0.00 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr5_+_135468516 | 0.00 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr9_+_32783497 | 0.00 |
ENST00000342743.5
|
TMEM215
|
transmembrane protein 215 |
| chr7_-_42971759 | 0.00 |
ENST00000538645.1
ENST00000445517.1 ENST00000223321.4 |
PSMA2
|
proteasome (prosome, macropain) subunit, alpha type, 2 |
| chr9_-_74383799 | 0.00 |
ENST00000377044.4
|
TMEM2
|
transmembrane protein 2 |
| chr11_+_64009072 | 0.00 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr5_-_140998616 | 0.00 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr14_-_54908043 | 0.00 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr12_-_31945172 | 0.00 |
ENST00000340398.3
|
H3F3C
|
H3 histone, family 3C |
| chr5_-_146833485 | 0.00 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
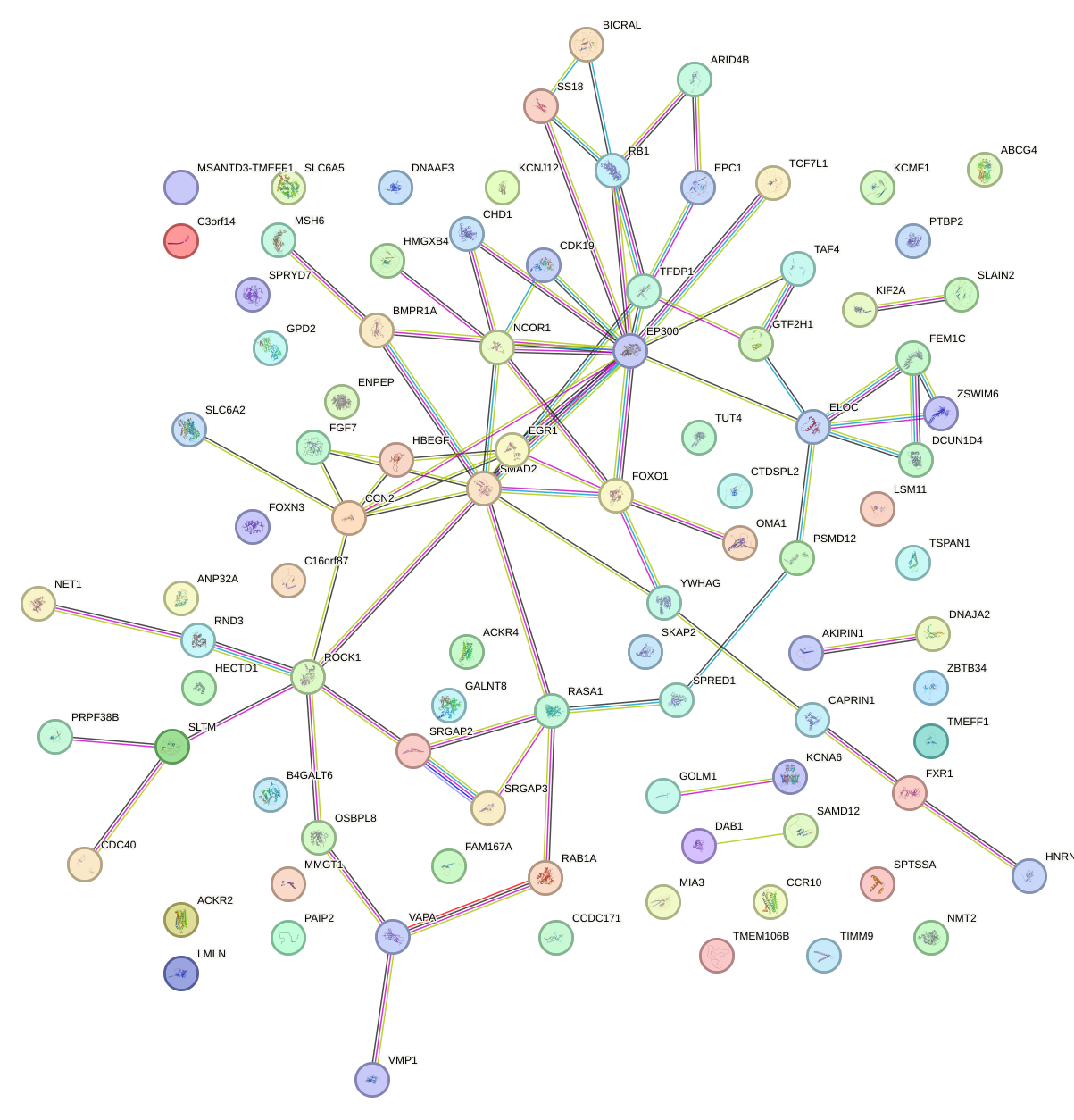
Network of associatons between targets according to the STRING database.
First level regulatory network of AACAGUC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) |
| 0.0 | 0.0 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |