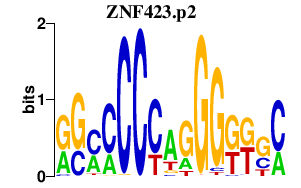
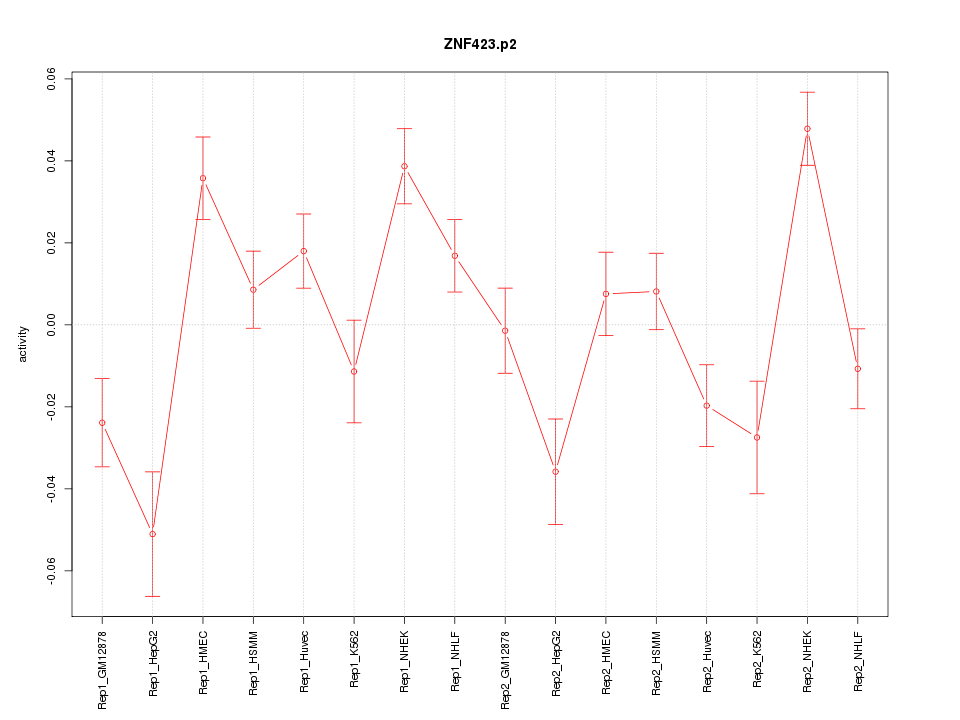
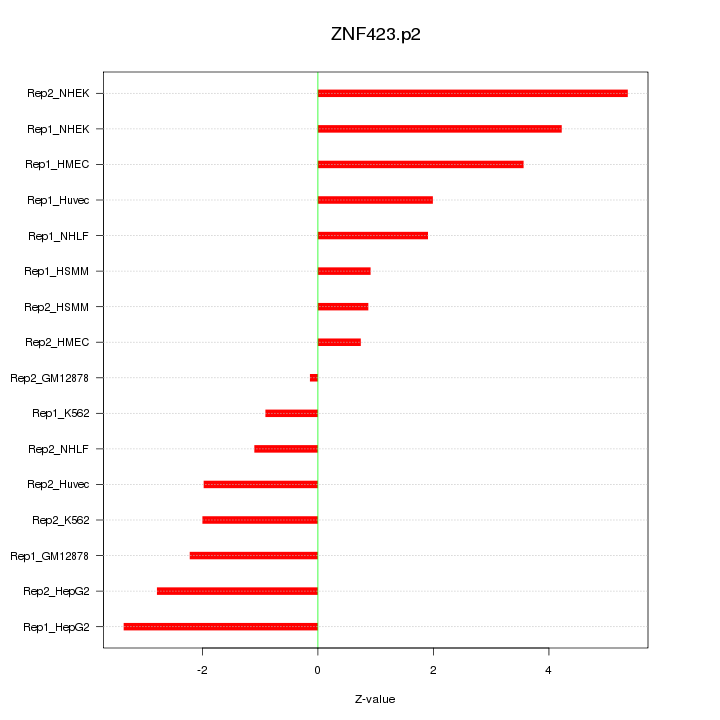
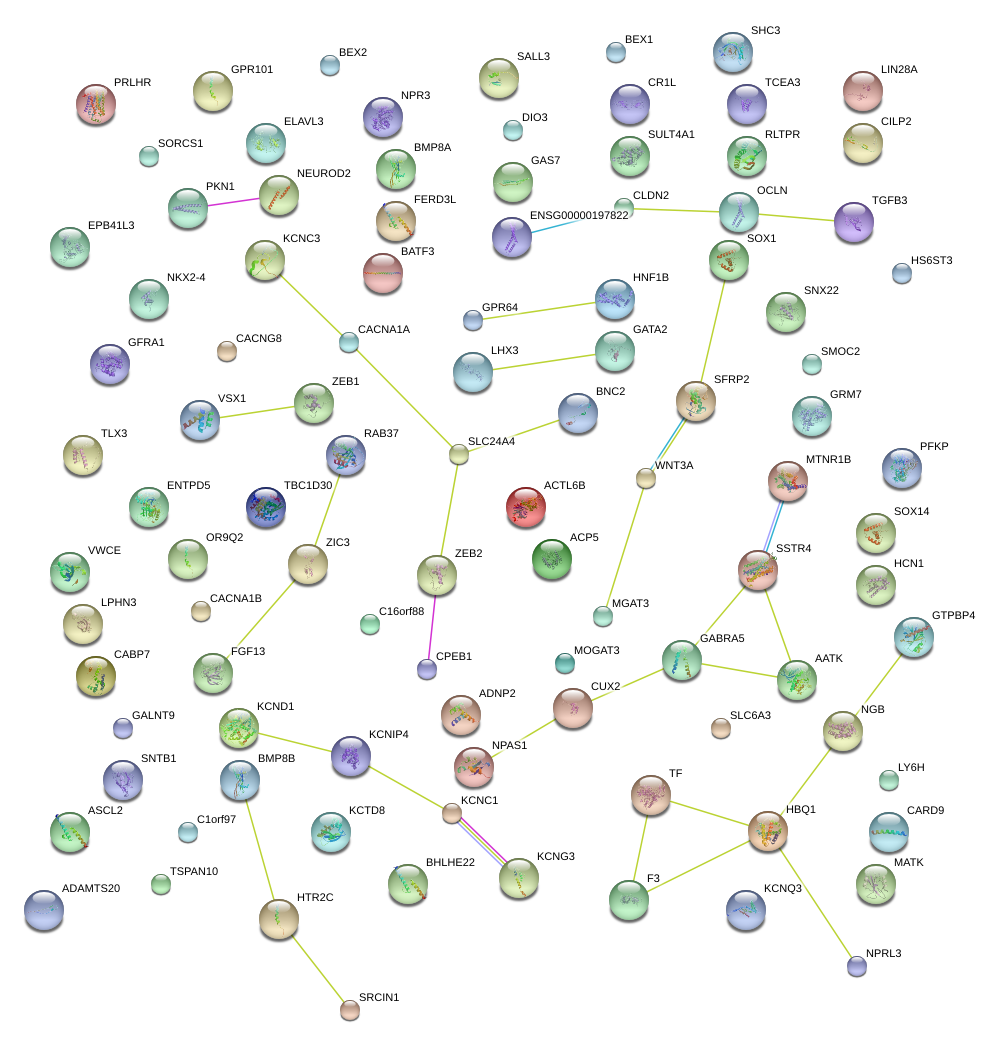
|
chr17_-_33178987
|
5.046
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr5_+_170668892
|
4.674
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr20_-_21326046
|
4.388
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr7_-_19151509
|
3.763
|
NM_152898
|
FERD3L
|
Fer3-like (Drosophila)
|
|
chr12_+_109956210
|
3.509
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr15_-_81113676
|
3.328
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr15_+_62230962
|
3.139
|
NM_024798
|
SNX22
|
sorting nexin 22
|
|
chr19_+_19510056
|
3.080
|
NM_153221
|
CILP2
|
cartilage intermediate layer protein 2
|
|
chr19_+_19510098
|
2.985
|
|
CILP2
|
cartilage intermediate layer protein 2
|
|
chr17_-_35017650
|
2.975
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr8_-_144313293
|
2.959
|
NM_001130478
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr19_+_14405100
|
2.887
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr22_+_38183480
|
2.739
|
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chrX_+_136475979
|
2.707
|
NM_003413
|
ZIC3
|
Zic family member 3 (odd-paired homolog, Drosophila)
|
|
chr17_-_35017691
|
2.553
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr3_+_6877812
|
2.533
|
|
GRM7
|
glutamate receptor, metabotropic 7
|
|
chr10_-_120345148
|
2.514
|
NM_004248
|
PRLHR
|
prolactin releasing hormone receptor
|
|
chr3_+_6877797
|
2.504
|
NM_000844
NM_181874
|
GRM7
|
glutamate receptor, metabotropic 7
|
|
chr19_-_55524445
|
2.465
|
NM_004977
|
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3
|
|
chr5_+_68823874
|
2.437
|
NM_002538
|
OCLN
|
occludin
|
|
chrX_-_19050499
|
2.420
|
NM_001079858
NM_001079859
NM_001079860
NM_001184833
NM_001184834
NM_001184835
NM_001184836
NM_001184837
NM_005756
|
GPR64
|
G protein-coupled receptor 64
|
|
chr3_+_134947940
|
2.401
|
|
TF
|
transferrin
|
|
chr4_+_61749540
|
2.399
|
|
LPHN3
|
latrophilin 3
|
|
chrX_-_138114100
|
2.387
|
|
FGF13
|
fibroblast growth factor 13
|
|
chr8_-_133561738
|
2.367
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chrX_+_136476353
|
2.364
|
|
ZIC3
|
Zic family member 3 (odd-paired homolog, Drosophila)
|
|
chr5_-_1498510
|
2.331
|
NM_001044
|
SLC6A3
|
solute carrier family 6 (neurotransmitter transporter, dopamine), member 3
|
|
chr12_-_42231990
|
2.329
|
NM_025003
|
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20
|
|
chr3_+_134947894
|
2.324
|
|
TF
|
transferrin
|
|
chr17_-_33179118
|
2.311
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr8_-_133561933
|
2.302
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chrX_-_48713194
|
2.301
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|
|
chr2_-_42574587
|
2.264
|
NM_133329
NM_172344
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr1_+_226261374
|
2.261
|
NM_033131
|
WNT3A
|
wingless-type MMTV integration site family, member 3A
|
|
chr4_-_44145266
|
2.207
|
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chr11_-_2248374
|
2.184
|
|
ASCL2
|
achaete-scute complex homolog 2 (Drosophila)
|
|
chr3_-_129694717
|
2.169
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr13_-_50315832
|
2.145
|
NM_198989
|
DLEU7
|
deleted in lymphocytic leukemia, 7
|
|
chr11_-_60819273
|
2.129
|
NM_152718
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr17_-_10042579
|
2.126
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr15_-_93671314
|
2.097
|
|
LOC400456
|
hypothetical LOC400456
|
|
chr6_+_168584678
|
2.091
|
NM_001166412
NM_022138
|
SMOC2
|
SPARC related modular calcium binding 2
|
|
chrX_+_113724806
|
2.071
|
NM_000868
|
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chr8_-_121892877
|
2.036
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr1_-_23623713
|
2.016
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr14_+_91859904
|
1.999
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr4_-_154929657
|
1.963
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|
|
chr20_+_22963949
|
1.944
|
NM_001052
|
SSTR4
|
somatostatin receptor 4
|
|
chr11_+_17714070
|
1.921
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr16_+_170334
|
1.921
|
NM_005331
|
HBQ1
|
hemoglobin, theta 1
|
|
chr10_-_118022965
|
1.915
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr4_-_20594645
|
1.905
|
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr5_+_68824143
|
1.892
|
|
OCLN
|
occludin
|
|
chr19_-_3737279
|
1.863
|
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr22_+_38183262
|
1.848
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr22_+_28446288
|
1.847
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr9_-_138234707
|
1.810
|
NM_014564
|
LHX3
|
LIM homeobox 3
|
|
chr19_+_59158101
|
1.798
|
NM_031895
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8
|
|
chr1_-_210939820
|
1.792
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr8_+_65656164
|
1.781
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr18_-_5533967
|
1.773
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr19_-_11550765
|
1.769
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr9_-_16860719
|
1.759
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr8_-_133562185
|
1.746
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr7_-_100091912
|
1.741
|
|
ACTL6B
|
actin-like 6B
|
|
chr6_+_168584722
|
1.738
|
|
SMOC2
|
SPARC related modular calcium binding 2
|
|
chr7_-_100092003
|
1.732
|
NM_016188
|
ACTL6B
|
actin-like 6B
|
|
chr22_-_42589543
|
1.716
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr10_+_3099708
|
1.706
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr1_+_205885080
|
1.686
|
NM_175710
|
CR1L
|
complement component (3b/4b) receptor 1-like
|
|
chr14_+_101097440
|
1.672
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chr19_+_14405216
|
1.663
|
|
PKN1
|
protein kinase N1
|
|
chr9_-_90983195
|
1.659
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr12_-_131415854
|
1.641
|
NM_001122636
|
GALNT9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9)
|
|
chr3_+_138965844
|
1.639
|
|
SOX14
|
SRY (sex determining region Y)-box 14
|
|
chr5_+_32747194
|
1.629
|
NM_000908
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr5_-_45731976
|
1.607
|
NM_021072
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1
|
|
chr17_-_76754466
|
1.604
|
NM_001080395
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr16_+_66236512
|
1.602
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr7_+_6622051
|
1.596
|
NM_017560
|
ZNF853
|
zinc finger protein 853
|
|
chr11_+_92342436
|
1.582
|
NM_005959
|
MTNR1B
|
melatonin receptor 1B
|
|
chr8_-_144313100
|
1.578
|
NM_001135655
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr6_+_168584706
|
1.572
|
|
SMOC2
|
SPARC related modular calcium binding 2
|
|
chr2_-_144991386
|
1.567
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_-_129690056
|
1.566
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr20_-_25010756
|
1.565
|
NM_014588
NM_199425
|
VSX1
|
visual system homeobox 1
|
|
chr18_+_75967902
|
1.560
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chrX_-_102205736
|
1.554
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr12_+_63504618
|
1.551
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr17_+_70244553
|
1.541
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr19_+_52214885
|
1.536
|
|
NPAS1
|
neuronal PAS domain protein 1
|
|
chr12_+_63504708
|
1.512
|
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr5_-_45731588
|
1.511
|
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1
|
|
chr1_-_40026718
|
1.505
|
|
BMP8B
|
bone morphogenetic protein 8b
|
|
chr1_+_26609893
|
1.504
|
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr19_-_11452439
|
1.503
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr13_+_95541093
|
1.485
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr10_-_108914277
|
1.463
|
NM_001013031
NM_052918
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chr17_-_34014390
|
1.462
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr14_-_76807302
|
1.456
|
NM_021257
|
NGB
|
neuroglobin
|
|
chr17_+_41432452
|
1.425
|
NM_001007532
|
STH
|
saitohin
|
|
chr9_+_139892049
|
1.410
|
NM_000718
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chrX_+_106050261
|
1.405
|
NM_020384
|
CLDN2
|
claudin 2
|
|
chr10_-_118022707
|
1.399
|
NM_001145453
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chrX_-_135941498
|
1.394
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr18_+_74841262
|
1.394
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr12_-_131256525
|
1.394
|
NM_021808
|
GALNT9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9)
|
|
chr19_-_3737221
|
1.394
|
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr17_+_70244945
|
1.392
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr15_+_24662958
|
1.386
|
NM_000810
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chrX_-_18282698
|
1.377
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr19_+_53589943
|
1.364
|
NM_000836
|
GRIN2D
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D
|
|
chr15_-_75711743
|
1.353
|
NM_032808
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr10_-_95350949
|
1.349
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr11_-_60819110
|
1.343
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr1_+_26609819
|
1.342
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr19_-_11550576
|
1.337
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr8_+_121892719
|
1.328
|
|
|
|
|
chr11_+_76561471
|
1.328
|
|
MYO7A
|
myosin VIIA
|
|
chr21_+_25856311
|
1.321
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr5_-_134397740
|
1.315
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr8_-_144313053
|
1.313
|
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr9_-_37024358
|
1.312
|
NM_016734
|
PAX5
|
paired box 5
|
|
chr11_+_14621844
|
1.311
|
NM_000922
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr11_-_14621738
|
1.304
|
NM_148976
|
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1
|
|
chr11_-_29995063
|
1.303
|
NM_002233
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4
|
|
chr9_+_134027154
|
1.301
|
NM_032536
|
NTNG2
|
netrin G2
|
|
chrX_-_101297417
|
1.294
|
NM_001159560
NM_001012978
|
BEX5
|
brain expressed, X-linked 5
|
|
chr15_+_24663199
|
1.291
|
NM_001165037
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5
|
|
chr18_-_42590989
|
1.265
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr11_+_14621914
|
1.253
|
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr16_-_27982277
|
1.245
|
NM_001109763
|
GSG1L
|
GSG1-like
|
|
chr10_+_134823925
|
1.242
|
NM_152643
|
KNDC1
|
kinase non-catalytic C-lobe domain (KIND) containing 1
|
|
chr17_+_45993338
|
1.238
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr11_+_14621884
|
1.234
|
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr21_+_25933637
|
1.209
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr20_-_49592444
|
1.208
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr19_-_55763091
|
1.207
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr12_-_48387394
|
1.205
|
|
FMNL3
|
formin-like 3
|
|
chr20_+_41978195
|
1.205
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr19_-_13478037
|
1.202
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr19_-_54005916
|
1.198
|
NM_001164773
NM_001190
|
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial
|
|
chr12_-_116021473
|
1.179
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chr15_+_82113813
|
1.173
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr21_+_25933916
|
1.173
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr7_+_103756339
|
1.160
|
NM_199000
|
LHFPL3
|
lipoma HMGIC fusion partner-like 3
|
|
chr17_+_73638430
|
1.160
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr3_-_58538110
|
1.154
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr16_+_53915971
|
1.150
|
NM_024335
|
IRX6
|
iroquois homeobox 6
|
|
chr16_+_57055717
|
1.148
|
|
NDRG4
|
NDRG family member 4
|
|
chr4_+_8633116
|
1.138
|
NM_080819
|
GPR78
|
G protein-coupled receptor 78
|
|
chr16_-_2186465
|
1.137
|
NM_020764
|
CASKIN1
|
CASK interacting protein 1
|
|
chr9_-_138236775
|
1.136
|
NM_178138
|
LHX3
|
LIM homeobox 3
|
|
chr15_-_77169894
|
1.132
|
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr21_+_25933938
|
1.129
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr17_+_19377732
|
1.121
|
NM_018242
|
SLC47A1
|
solute carrier family 47, member 1
|
|
chr12_-_48387459
|
1.119
|
NM_175736
NM_198900
|
FMNL3
|
formin-like 3
|
|
chr6_+_111515341
|
1.111
|
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chrX_+_152562013
|
1.110
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr15_-_75711785
|
1.108
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr5_-_136862786
|
1.104
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr16_+_66236559
|
1.103
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr15_+_77362200
|
1.096
|
NM_001146341
|
ANKRD34C
|
ankyrin repeat domain 34C
|
|
chr1_-_210939727
|
1.092
|
|
|
|
|
chr19_-_5291700
|
1.091
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr11_-_132318080
|
1.087
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr18_+_2896178
|
1.086
|
|
EMILIN2
|
elastin microfibril interfacer 2
|
|
chr16_-_87297525
|
1.084
|
NM_001171816
|
RNF166
|
ring finger protein 166
|
|
chr7_-_71439678
|
1.079
|
|
CALN1
|
calneuron 1
|
|
chr6_+_39868126
|
1.074
|
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr5_-_136862317
|
1.072
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr3_+_123385870
|
1.072
|
NM_001178065
|
CASR
|
calcium-sensing receptor
|
|
chr1_-_38285033
|
1.070
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr1_-_210939929
|
1.067
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr14_+_103674812
|
1.064
|
NM_015656
|
KIF26A
|
kinesin family member 26A
|
|
chr9_+_137107222
|
1.059
|
|
OLFM1
|
olfactomedin 1
|
|
chr9_-_23811842
|
1.053
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr12_-_48387233
|
1.051
|
|
|
|
|
chr3_+_185430910
|
1.049
|
NM_138345
|
VWA5B2
|
von Willebrand factor A domain containing 5B2
|
|
chr3_-_13436725
|
1.039
|
|
NUP210
|
nucleoporin 210kDa
|
|
chr10_+_135190855
|
1.039
|
NM_000773
|
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1
|
|
chr17_+_50698169
|
1.037
|
|
HLF
|
hepatic leukemia factor
|
|
chr11_-_117173004
|
1.034
|
|
DSCAML1
|
Down syndrome cell adhesion molecule like 1
|
|
chr19_-_3737382
|
1.031
|
NM_139354
NM_139355
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr4_-_44145580
|
1.029
|
NM_198353
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chr3_+_37468542
|
1.027
|
NM_002207
|
ITGA9
|
integrin, alpha 9
|
|
chr21_+_45699830
|
1.027
|
NM_030582
NM_130444
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr6_+_39868116
|
1.021
|
|
|
|
|
chr12_-_63801539
|
1.013
|
|
WIF1
|
WNT inhibitory factor 1
|
|
chr4_-_46086528
|
1.008
|
NM_000807
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2
|
|
chr7_-_156495985
|
1.007
|
NM_005515
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr18_+_11741470
|
0.996
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chrX_+_30175191
|
0.988
|
NM_177404
|
MAGEB1
|
melanoma antigen family B, 1
|
|
chrX_+_151975518
|
0.986
|
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr1_+_16247870
|
0.984
|
NM_001165945
|
CLCNKB
|
chloride channel Kb
|
|
chr20_-_42872295
|
0.980
|
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr3_-_124228999
|
0.960
|
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B
|
|
chrX_+_152413646
|
0.958
|
|
BGN
|
biglycan
|
|
chr17_+_8865547
|
0.951
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr16_-_3131348
|
0.945
|
|
|
|