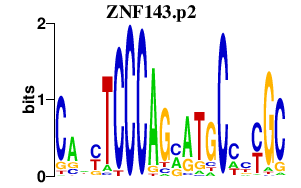
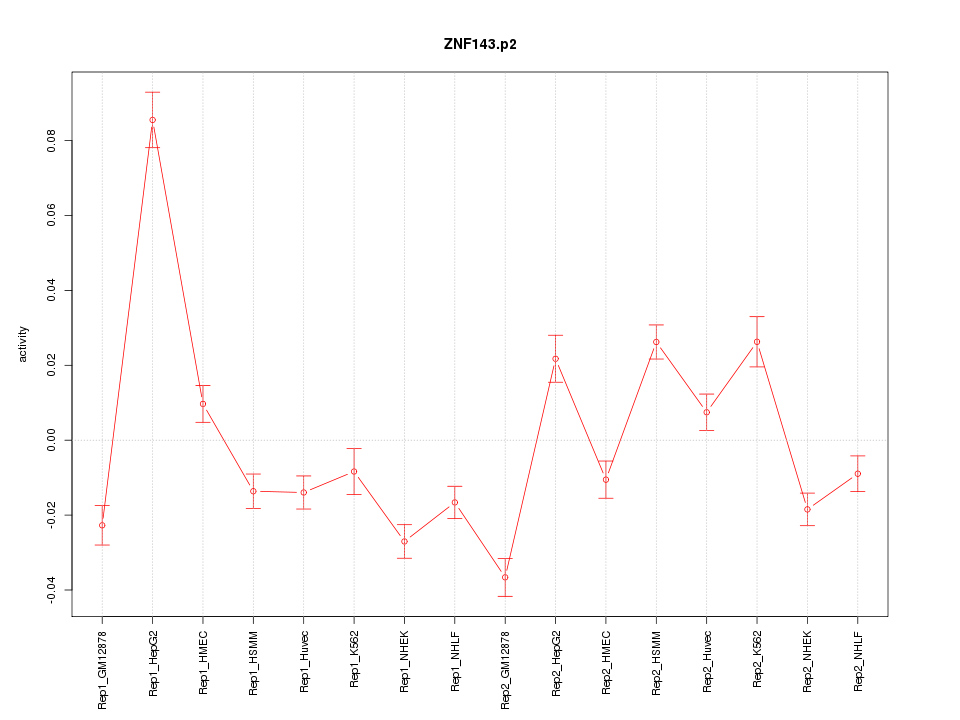
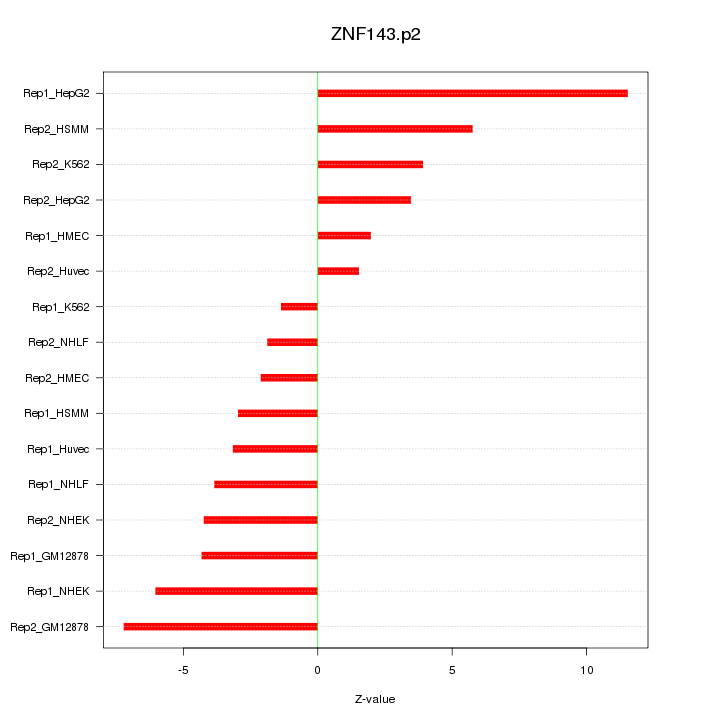
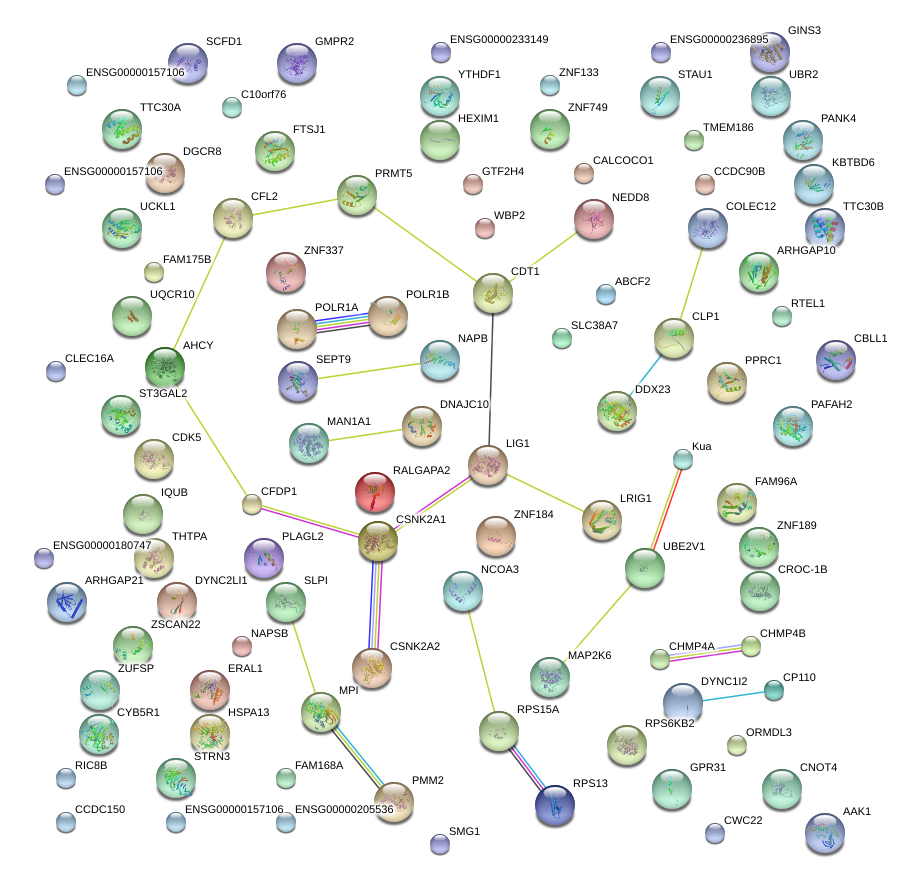
|
chr20_-_62058162
|
9.196
|
NM_017859
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr16_-_30841938
|
8.892
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr20_-_32354736
|
7.379
|
NM_000687
|
AHCY
|
adenosylhomocysteinase
|
|
chr22_+_28493351
|
5.593
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr2_-_178125680
|
5.436
|
NM_152517
|
TTC30B
|
tetratricopeptide repeat domain 30B
|
|
chr7_-_134845363
|
5.105
|
NM_001008225
NM_001190847
NM_001190848
NM_001190849
NM_001190850
NM_013316
|
CNOT4
|
CCR4-NOT transcription complex, subunit 4
|
|
chr20_+_31862789
|
5.026
|
|
CHMP4B
|
chromatin modifying protein 4B
|
|
chr6_-_27548438
|
4.935
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr10_-_42368035
|
4.875
|
|
ZNF37BP
|
zinc finger protein 37B, pseudogene
|
|
chr12_+_105692466
|
4.786
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr19_+_63530155
|
4.752
|
NM_181846
|
ZSCAN22
|
zinc finger and SCAN domain containing 22
|
|
chr16_+_8799170
|
4.661
|
NM_000303
|
PMM2
|
phosphomannomutase 2
|
|
chr20_-_23350038
|
4.554
|
NM_022080
|
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta
|
|
chr2_-_86186441
|
4.540
|
|
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa
|
|
chr12_-_47532171
|
4.440
|
NM_004818
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr1_+_154291140
|
4.374
|
NM_001145264
NM_014017
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2
|
|
chr2_+_172252473
|
4.368
|
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2
|
|
chr6_+_42639929
|
4.299
|
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr20_+_61760084
|
4.258
|
NM_016434
NM_032957
|
RTEL1
|
regulator of telomere elongation helicase 1
|
|
chr14_-_23095240
|
4.234
|
|
|
|
|
chr17_-_35337288
|
4.228
|
NM_139280
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr19_-_19748186
|
3.960
|
|
LOC284440
|
hypothetical LOC284440
|
|
chr11_-_72986738
|
3.948
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr14_-_22468393
|
3.944
|
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr16_-_8798973
|
3.933
|
NM_015421
|
TMEM186
|
transmembrane protein 186
|
|
chr11_-_17055763
|
3.930
|
NM_001017
|
RPS13
|
ribosomal protein S13
|
|
chr16_-_69030421
|
3.915
|
NM_006927
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr16_+_19442679
|
3.901
|
NM_001199022
NM_014711
|
CP110
|
CP110 protein
|
|
chr11_+_57181791
|
3.775
|
NM_001142597
NM_006831
|
CLP1
|
CLP1, cleavage and polyadenylation factor I subunit, homolog (S. cerevisiae)
|
|
chr14_+_23095028
|
3.654
|
NM_001126339
NM_024328
|
THTPA
|
thiamine triphosphatase
|
|
chr9_+_103200939
|
3.585
|
NM_003452
NM_197977
|
ZNF189
|
zinc finger protein 189
|
|
chr6_+_11202251
|
3.581
|
NM_001135575
|
LOC221710
|
hypothetical protein LOC221710
|
|
chr6_+_11201977
|
3.575
|
|
LOC221710
|
hypothetical protein LOC221710
|
|
chr11_+_66952462
|
3.561
|
NM_003952
|
RPS6KB2
|
ribosomal protein S6 kinase, 70kDa, polypeptide 2
|
|
chr20_-_472335
|
3.540
|
NM_001895
NM_177559
NM_177560
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide
|
|
chr15_+_72969404
|
3.482
|
NM_002435
|
MPI
|
mannose phosphate isomerase
|
|
chr6_-_117096594
|
3.469
|
NM_145062
|
ZUFSP
|
zinc finger with UFM1-specific peptidase domain
|
|
chr16_+_56983798
|
3.449
|
NM_001126129
NM_001126130
NM_022770
|
GINS3
|
GINS complex subunit 3 (Psf3 homolog)
|
|
chr14_+_23771464
|
3.384
|
NM_001002000
NM_001002001
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr2_-_180580024
|
3.364
|
NM_020943
|
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chr11_-_82674546
|
3.362
|
|
CCDC90B
|
coiled-coil domain containing 90B
|
|
chr15_-_62173041
|
3.321
|
NM_001014812
NM_032231
|
FAM96A
|
family with sequence similarity 96, member A
|
|
chr16_-_18845223
|
3.318
|
NM_015092
|
SMG1
|
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chr14_+_23771942
|
3.296
|
NM_001002002
NM_016576
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr14_-_22468445
|
3.276
|
NM_001039619
NM_006109
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr6_-_119712596
|
3.264
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr16_-_18709125
|
3.213
|
NM_001019
NM_001030009
|
RPS15A
|
ribosomal protein S15a
|
|
chr20_-_30259007
|
3.206
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr17_+_24206100
|
3.135
|
NM_005702
|
ERAL1
|
Era G-protein-like 1 (E. coli)
|
|
chr20_+_18217120
|
3.098
|
NM_001083330
NM_003434
|
ZNF133
|
zinc finger protein 133
|
|
chr17_+_64922420
|
3.093
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chrX_+_48219344
|
3.066
|
NM_012280
NM_177439
|
FTSJ1
|
FtsJ homolog 1 (E. coli)
|
|
chr1_-_2447848
|
3.009
|
NM_018216
|
PANK4
|
pantothenate kinase 4
|
|
chr20_+_45564063
|
3.002
|
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr10_+_103882776
|
2.981
|
NM_015062
|
PPRC1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1
|
|
chr12_-_52407470
|
2.978
|
NM_001143682
NM_020898
|
CALCOCO1
|
calcium binding and coiled-coil domain 1
|
|
chr14_-_23771399
|
2.970
|
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr7_-_150385912
|
2.942
|
NM_001164410
NM_004935
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr2_+_43854681
|
2.937
|
NM_001193464
NM_015522
NM_016008
|
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1
|
|
chr21_-_14677310
|
2.930
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr2_+_197212522
|
2.876
|
NM_001080539
|
CCDC150
|
coiled-coil domain containing 150
|
|
chr2_-_178191618
|
2.864
|
|
TTC30A
|
tetratricopeptide repeat domain 30A
|
|
chr20_-_61317891
|
2.851
|
NM_017798
|
YTHDF1
|
YTH domain family, member 1
|
|
chr19_-_53365371
|
2.838
|
NM_000234
|
LIG1
|
ligase I, DNA, ATP-dependent
|
|
chr22_+_18447732
|
2.778
|
NM_001190326
NM_022720
|
DGCR8
|
DiGeorge syndrome critical region gene 8
|
|
chr14_+_30161285
|
2.760
|
|
SCFD1
|
sec1 family domain containing 1
|
|
chr20_-_48165886
|
2.755
|
NM_022442
NM_021988
NM_199144
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr19_-_41711009
|
2.748
|
NM_001012756
NM_001166036
NM_001166037
NM_001166038
|
ZNF260
|
zinc finger protein 260
|
|
chr16_-_74024750
|
2.744
|
NM_006324
|
CFDP1
|
craniofacial development protein 1
|
|
chr2_+_183289205
|
2.713
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr19_+_62638474
|
2.697
|
NM_001023561
|
ZNF749
|
zinc finger protein 749
|
|
chr14_-_30565275
|
2.689
|
NM_001083893
NM_014574
|
STRN3
|
striatin, calmodulin binding protein 3
|
|
chr17_-_71362971
|
2.679
|
NM_012478
|
WBP2
|
WW domain binding protein 2
|
|
chr6_+_42639745
|
2.672
|
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr1_-_201202956
|
2.629
|
NM_016243
|
CYB5R1
|
cytochrome b5 reductase 1
|
|
chr20_-_47238082
|
2.594
|
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr20_-_20641122
|
2.591
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr7_+_107171809
|
2.581
|
|
CBLL1
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1
|
|
chr14_-_34253473
|
2.539
|
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr6_-_167491305
|
2.523
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr16_-_57276108
|
2.507
|
NM_018231
|
SLC38A7
|
solute carrier family 38, member 7
|
|
chr20_+_18496068
|
2.507
|
|
LOC388789
|
hypothetical LOC388789
|
|
chr13_-_40604587
|
2.491
|
NM_152903
|
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6
|
|
chr16_+_10945815
|
2.455
|
NM_015226
|
CLEC16A
|
C-type lectin domain family 16, member A
|
|
chr6_+_42639714
|
2.454
|
NM_001184801
NM_015255
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr10_-_103805858
|
2.442
|
NM_024541
|
C10orf76
|
chromosome 10 open reading frame 76
|
|
chr6_+_30983941
|
2.427
|
NM_001517
|
GTF2H4
|
general transcription factor IIH, polypeptide 4, 52kDa
|
|
chr14_-_23771312
|
2.421
|
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr20_-_25625173
|
2.420
|
NM_015655
|
ZNF337
|
zinc finger protein 337
|
|
chr1_-_26197121
|
2.408
|
NM_000437
|
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa
|
|
chr7_-_150555153
|
2.407
|
NM_005692
NM_007189
|
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2
|
|
chr14_-_23771401
|
2.407
|
NM_006156
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr4_+_148872902
|
2.404
|
NM_024605
|
ARHGAP10
|
Rho GTPase activating protein 10
|
|
chr10_+_126480343
|
2.388
|
NM_032182
|
FAM175B
|
family with sequence similarity 175, member B
|
|
chr20_+_45564007
|
2.371
|
NM_001174087
NM_001174088
NM_006534
NM_181659
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr2_-_69724480
|
2.363
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr11_-_82675024
|
2.338
|
NM_021825
|
CCDC90B
|
coiled-coil domain containing 90B
|
|
chr2_+_113016236
|
2.316
|
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa
|
|
chr20_-_18425811
|
2.309
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr7_-_150385867
|
2.296
|
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr3_+_98966253
|
2.293
|
NM_032146
NM_177976
|
ARL6
|
ADP-ribosylation factor-like 6
|
|
chr22_-_28492990
|
2.291
|
|
|
|
|
chr11_+_43622348
|
2.284
|
|
|
|
|
chr4_-_170167962
|
2.280
|
NM_032783
|
CBR4
|
carbonyl reductase 4
|
|
chr2_-_24123726
|
2.275
|
NM_001142319
NM_025203
|
C2orf44
|
chromosome 2 open reading frame 44
|
|
chr14_+_30161267
|
2.275
|
NM_016106
NM_182835
|
SCFD1
|
sec1 family domain containing 1
|
|
chr17_+_71292275
|
2.274
|
NM_001080419
|
UNK
|
unkempt homolog (Drosophila)
|
|
chr1_+_200584421
|
2.263
|
NM_001167857
NM_001167858
NM_002481
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr6_-_30818198
|
2.236
|
NM_005803
|
FLOT1
|
flotillin 1
|
|
chr2_-_20413838
|
2.235
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr4_-_47611255
|
2.216
|
NM_152995
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1
|
|
chrX_+_154764136
|
2.198
|
NM_001145149
NM_001185183
NM_005638
|
VAMP7
|
vesicle-associated membrane protein 7
|
|
chr2_-_148494732
|
2.189
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr2_-_55699489
|
2.188
|
|
SMEK2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium)
|
|
chr6_+_139136349
|
2.173
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr6_+_43705254
|
2.168
|
NM_001003690
|
MAD2L1BP
|
MAD2L1 binding protein
|
|
chr14_-_89868006
|
2.164
|
NM_017970
NM_199043
|
C14orf102
|
chromosome 14 open reading frame 102
|
|
chr2_+_95194903
|
2.163
|
NM_001017396
NM_021088
|
ZNF2
|
zinc finger protein 2
|
|
chr20_-_19981005
|
2.160
|
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr2_+_241174803
|
2.152
|
NM_023083
NM_023085
|
CAPN10
|
calpain 10
|
|
chr7_+_111877699
|
2.149
|
NM_001550
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr19_-_42962039
|
2.142
|
NM_001172689
NM_001172690
NM_001172691
NM_001172692
NM_152360
|
ZNF573
|
zinc finger protein 573
|
|
chr20_-_13713509
|
2.136
|
NM_016649
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae)
|
|
chr2_+_95195260
|
2.132
|
|
ZNF2
|
zinc finger protein 2
|
|
chr12_+_56168082
|
2.125
|
NM_004990
|
MARS
|
methionyl-tRNA synthetase
|
|
chr2_+_232354675
|
2.122
|
|
COPS7B
|
COP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis)
|
|
chr2_-_88707978
|
2.121
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr7_+_134893185
|
2.102
|
NM_015135
|
NUP205
|
nucleoporin 205kDa
|
|
chr17_+_40494167
|
2.102
|
|
NMT1
|
N-myristoyltransferase 1
|
|
chr2_-_86186788
|
2.098
|
NM_015425
|
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa
|
|
chr13_-_32010883
|
2.095
|
NM_014887
NM_033111
|
N4BP2L2
|
NEDD4 binding protein 2-like 2
|
|
chr14_+_35365266
|
2.093
|
NM_032352
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like
|
|
chr22_+_25155220
|
2.076
|
NM_020437
|
ASPHD2
|
aspartate beta-hydroxylase domain containing 2
|
|
chr16_-_66902286
|
2.075
|
NM_032178
|
SLC7A6OS
|
solute carrier family 7, member 6 opposite strand
|
|
chr17_+_38576707
|
2.064
|
NM_005899
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr6_-_84994032
|
2.054
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr16_+_66902369
|
2.045
|
NM_001184824
NM_019023
|
PRMT7
|
protein arginine methyltransferase 7
|
|
chr6_-_17814596
|
2.045
|
NM_005124
|
NUP153
|
nucleoporin 153kDa
|
|
chr20_+_31862707
|
2.045
|
NM_176812
|
CHMP4B
|
chromatin modifying protein 4B
|
|
chr10_+_75211810
|
2.029
|
NM_203298
|
CHCHD1
|
coiled-coil-helix-coiled-coil-helix domain containing 1
|
|
chr2_-_176575171
|
2.023
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chr12_-_62902279
|
2.017
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr2_-_27446816
|
2.014
|
NM_001034116
NM_015636
|
EIF2B4
|
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa
|
|
chr1_-_45924770
|
2.000
|
NM_021639
|
GPBP1L1
|
GC-rich promoter binding protein 1-like 1
|
|
chr17_+_44263370
|
1.994
|
NM_005831
|
CALCOCO2
|
calcium binding and coiled-coil domain 2
|
|
chr8_+_38877909
|
1.992
|
NM_021623
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2
|
|
chr11_-_77577288
|
1.979
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr12_+_56168120
|
1.964
|
|
MARS
|
methionyl-tRNA synthetase
|
|
chr19_+_62483606
|
1.963
|
NM_006635
|
ZNF460
|
zinc finger protein 460
|
|
chr2_+_171494026
|
1.956
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr17_-_7173179
|
1.946
|
NM_001005408
NM_032442
|
NEURL4
|
neuralized homolog 4 (Drosophila)
|
|
chr6_+_79633907
|
1.945
|
NM_001010844
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1
|
|
chr3_-_168935287
|
1.944
|
NM_007217
NM_145860
NM_145859
|
PDCD10
|
programmed cell death 10
|
|
chr3_-_172660545
|
1.935
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr16_+_56567839
|
1.933
|
NM_199046
NM_199456
|
TEPP
|
testis, prostate and placenta expressed
|
|
chr6_+_108722752
|
1.917
|
NM_145315
|
LACE1
|
lactation elevated 1
|
|
chr19_-_47438583
|
1.915
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr2_+_172252194
|
1.904
|
NM_001378
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2
|
|
chr16_-_23515104
|
1.902
|
NM_005003
|
NDUFAB1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa
|
|
chr3_+_101462375
|
1.901
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr16_+_4415805
|
1.895
|
NM_001135110
NM_005147
|
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3
|
|
chr11_-_6661093
|
1.889
|
NM_022061
|
MRPL17
|
mitochondrial ribosomal protein L17
|
|
chr19_-_47438563
|
1.885
|
NM_019884
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr3_+_102926126
|
1.872
|
NM_024548
|
CEP97
|
centrosomal protein 97kDa
|
|
chr20_+_762355
|
1.869
|
NM_001042353
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr6_+_109523388
|
1.863
|
|
CEP57L1
|
centrosomal protein 57kDa-like 1
|
|
chr1_-_142309117
|
1.862
|
|
|
|
|
chr3_-_130600931
|
1.860
|
|
RPL32P3
|
ribosomal protein L32 pseudogene 3
|
|
chr2_+_32244406
|
1.854
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr20_-_44575599
|
1.845
|
NM_018102
NM_199441
|
ZNF334
|
zinc finger protein 334
|
|
chr15_-_47235057
|
1.843
|
NM_001143887
NM_004236
|
COPS2
|
COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis)
|
|
chr2_-_178191923
|
1.833
|
NM_152275
|
TTC30A
|
tetratricopeptide repeat domain 30A
|
|
chr12_-_94953427
|
1.830
|
NM_000895
|
LTA4H
|
leukotriene A4 hydrolase
|
|
chr20_+_1254033
|
1.814
|
|
LOC100507495
|
hypothetical LOC100507495
|
|
chr16_+_69045998
|
1.813
|
NM_145059
|
FUK
|
fucokinase
|
|
chr16_-_626293
|
1.813
|
NM_001040160
NM_001040161
NM_001040162
NM_001040165
NM_032366
|
C16orf13
|
chromosome 16 open reading frame 13
|
|
chr19_-_47438548
|
1.813
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr17_+_40494201
|
1.812
|
NM_021079
|
NMT1
|
N-myristoyltransferase 1
|
|
chr2_-_170138558
|
1.796
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr2_-_18605203
|
1.795
|
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis)
|
|
chr20_+_30259235
|
1.795
|
NM_015352
NM_172236
|
POFUT1
|
protein O-fucosyltransferase 1
|
|
chr5_+_85949518
|
1.794
|
NM_001867
|
COX7C
|
cytochrome c oxidase subunit VIIc
|
|
chr1_-_95311031
|
1.790
|
NM_144988
|
ALG14
|
asparagine-linked glycosylation 14 homolog (S. cerevisiae)
|
|
chr5_+_85949511
|
1.784
|
|
COX7C
|
cytochrome c oxidase subunit VIIc
|
|
chr2_-_234317362
|
1.782
|
NM_001001394
|
DNAJB3
|
DnaJ (Hsp40) homolog, subfamily B, member 3
|
|
chr17_+_16283025
|
1.782
|
|
NCRNA00188
|
non-protein coding RNA 188
|
|
chr20_-_32728517
|
1.780
|
|
PIGU
|
phosphatidylinositol glycan anchor biosynthesis, class U
|
|
chr12_-_119938593
|
1.768
|
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr22_-_28492933
|
1.764
|
NM_001003692
NM_019103
|
ZMAT5
|
zinc finger, matrin-type 5
|
|
chr2_+_75727415
|
1.763
|
NM_014763
|
MRPL19
|
mitochondrial ribosomal protein L19
|
|
chr20_+_13713645
|
1.759
|
NM_001039375
NM_024120
|
C20orf7
|
chromosome 20 open reading frame 7
|
|
chr19_+_3136965
|
1.759
|
|
NCLN
|
nicalin
|
|
chr2_-_202024317
|
1.758
|
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr6_-_43704742
|
1.751
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr17_-_17435631
|
1.737
|
NM_148172
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr2_+_53867571
|
1.735
|
NM_001127397
NM_001127398
NM_015701
|
ERLEC1
|
endoplasmic reticulum lectin 1
|
|
chr2_+_38746742
|
1.723
|
|
GALM
|
galactose mutarotase (aldose 1-epimerase)
|
|
chr19_-_49131250
|
1.707
|
NM_003425
|
ZNF45
|
zinc finger protein 45
|
|
chr7_+_7188667
|
1.707
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr20_-_32163576
|
1.706
|
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa
|