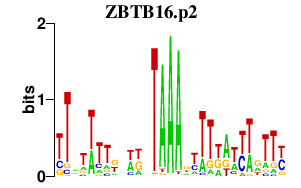
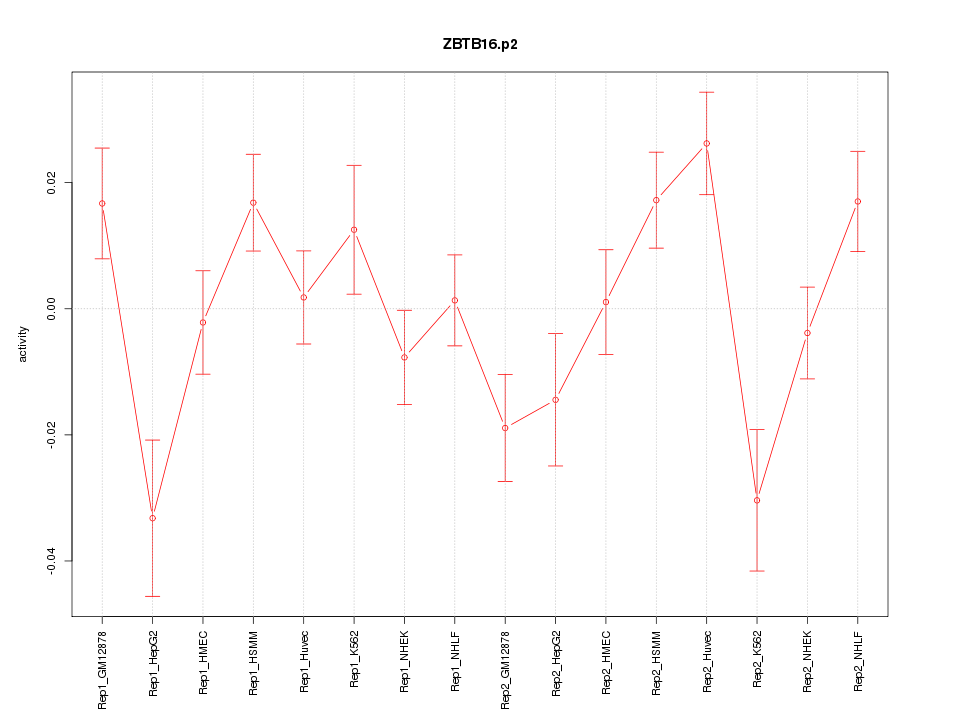
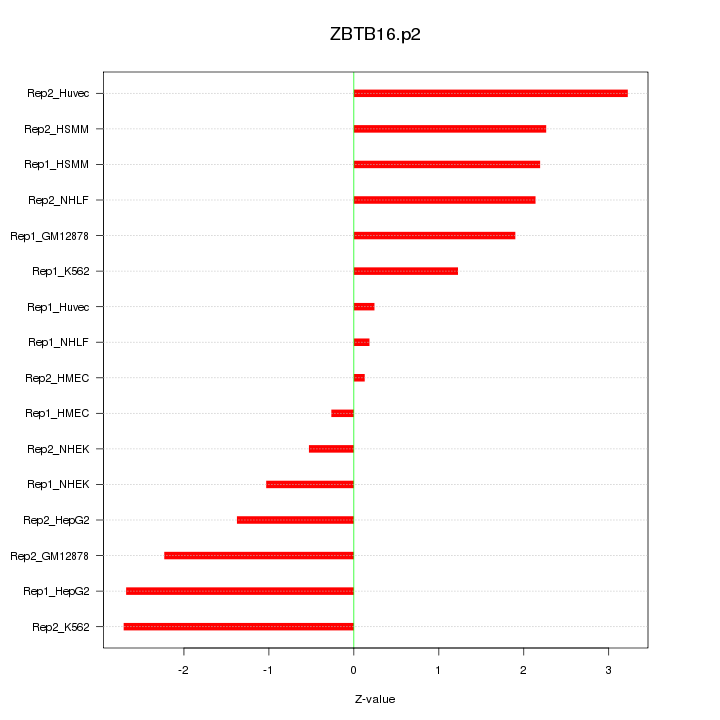
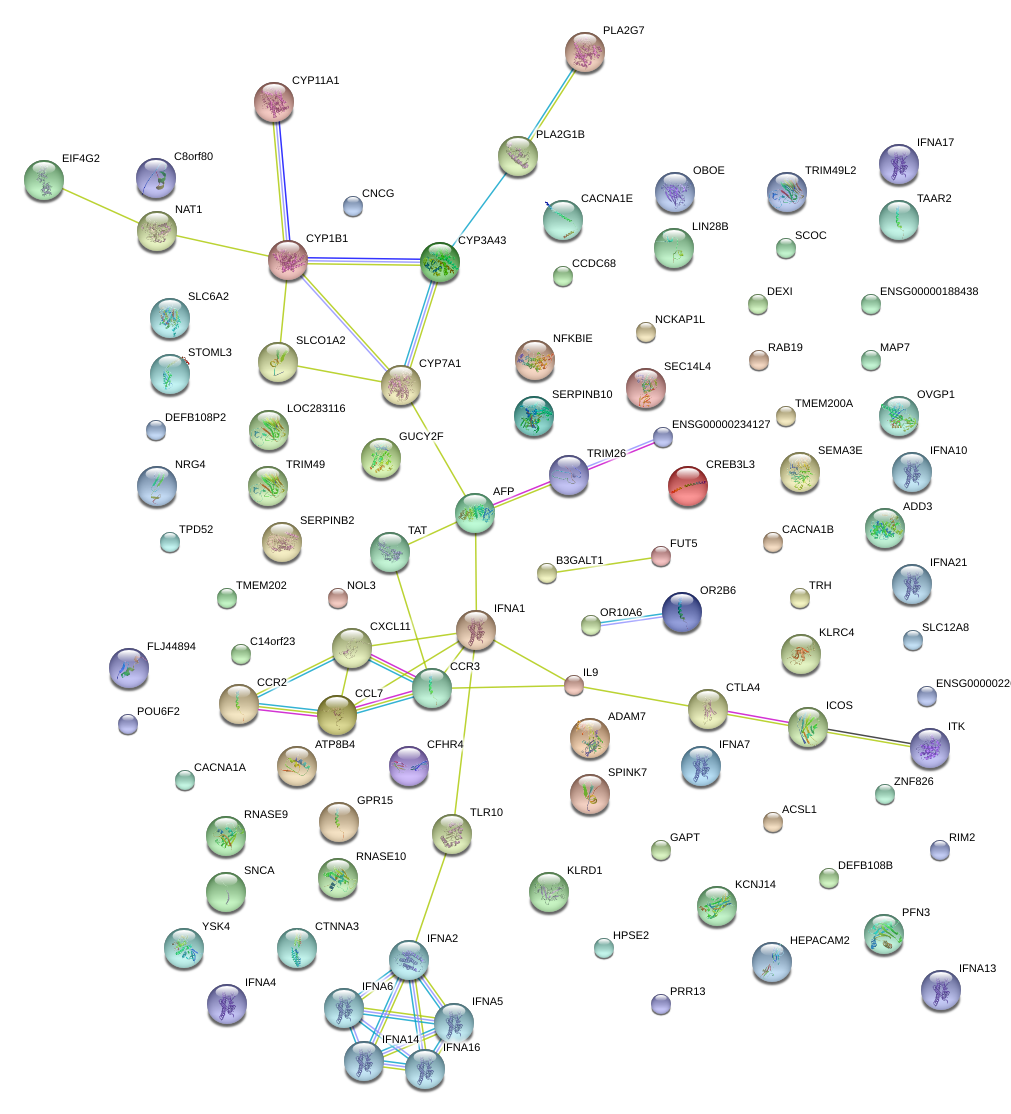
|
chr12_-_10453409
|
2.848
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 read-through transcript
killer cell lectin-like receptor subfamily C, member 4
|
|
chr5_+_57823017
|
2.779
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr9_-_21340885
|
2.753
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr4_-_77176163
|
2.645
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr7_-_92693716
|
2.547
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr4_+_74520796
|
2.290
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr4_-_38460983
|
2.114
|
NM_001017388
NM_001195106
NM_001195107
NM_001195108
NM_030956
|
TLR10
|
toll-like receptor 10
|
|
chr9_-_21375258
|
2.090
|
NM_000605
|
IFNA2
|
interferon, alpha 2
|
|
chr18_+_59733724
|
2.089
|
NM_005024
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr12_-_9987277
|
2.071
|
|
LOC100506159
|
hypothetical LOC100506159
|
|
chr7_+_99263571
|
2.039
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr5_-_176760242
|
1.999
|
NM_001029886
|
PFN3
|
profilin 3
|
|
chr2_-_135498716
|
1.953
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chr16_-_70168449
|
1.897
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr14_+_28311740
|
1.869
|
|
C14orf23
|
chromosome 14 open reading frame 23
|
|
chr17_+_29621352
|
1.849
|
NM_006273
|
CCL7
|
chemokine (C-C motif) ligand 7
|
|
chr4_-_90978469
|
1.839
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr12_+_10351683
|
1.831
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr9_+_21430439
|
1.760
|
NM_024013
|
IFNA1
|
interferon, alpha 1
|
|
chr15_-_72446979
|
1.644
|
NM_000781
|
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1
|
|
chr9_-_21218132
|
1.638
|
NM_021268
|
IFNA17
|
interferon, alpha 17
|
|
chr8_-_27997306
|
1.542
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr2_+_168383427
|
1.537
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr9_-_21295311
|
1.496
|
NM_002169
|
IFNA5
|
interferon, alpha 5
|
|
chr18_-_50777634
|
1.478
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr7_+_38984133
|
1.475
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr3_+_46370228
|
1.464
|
NM_001123041
NM_001123396
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr9_-_21229975
|
1.460
|
NM_002172
|
IFNA14
|
interferon, alpha 14
|
|
chr7_-_83108682
|
1.449
|
NM_001178129
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr4_+_141484046
|
1.449
|
NM_001153446
NM_001153635
NM_001153585
NM_001153690
|
SCOC
|
short coiled-coil protein
|
|
chr8_+_104900591
|
1.427
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr5_-_135259403
|
1.416
|
NM_000590
|
IL9
|
interleukin 9
|
|
chr8_-_59575249
|
1.400
|
NM_000780
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1
|
|
chr9_-_21177529
|
1.354
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr1_+_195123766
|
1.338
|
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr19_-_5821546
|
1.290
|
NM_002034
|
FUT5
|
fucosyltransferase 5 (alpha (1,3) fucosyltransferase)
|
|
chr7_-_83116414
|
1.270
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr10_-_100985548
|
1.270
|
NM_001166244
NM_001166245
NM_001166246
NM_021828
|
HPSE2
|
heparanase 2
|
|
chr15_+_26832300
|
1.240
|
|
LOC100289656
|
Dexi homolog (mouse) pseudogene
|
|
chr22_-_29231597
|
1.225
|
NM_001161368
NM_174977
|
SEC14L4
|
SEC14-like 4 (S. cerevisiae)
|
|
chr4_-_47709717
|
1.222
|
NM_000087
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr5_+_156540425
|
1.210
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr2_+_204509698
|
1.199
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr11_-_89181390
|
1.188
|
NM_020358
|
TRIM49
|
tripartite motif containing 49
|
|
chr19_-_13478037
|
1.172
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr1_-_111771877
|
1.171
|
NM_002557
|
OVGP1
|
oviductal glycoprotein 1, 120kDa
|
|
chr9_-_21156609
|
1.158
|
NM_002175
|
IFNA21
|
interferon, alpha 21
|
|
chr8_+_24354387
|
1.155
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr10_+_111755715
|
1.151
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr19_-_20399601
|
1.147
|
|
ZNF826P
|
zinc finger protein 826, pseudogene
|
|
chr6_+_130799954
|
1.141
|
NM_052913
|
TMEM200A
|
transmembrane protein 200A
|
|
chr9_-_21192201
|
1.139
|
NM_021057
|
IFNA7
|
interferon, alpha 7
|
|
chr5_-_88004891
|
1.139
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr12_+_63510481
|
1.138
|
|
|
|
|
chr15_-_74091817
|
1.134
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr2_-_38156769
|
1.119
|
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr19_+_53656317
|
1.119
|
NM_170720
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr17_+_23818922
|
1.116
|
|
|
|
|
chr12_-_21379098
|
1.115
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr9_-_21207303
|
1.112
|
NM_002173
|
IFNA16
|
interferon, alpha 16
|
|
chr6_-_46811388
|
1.105
|
NM_001168357
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr4_-_185963859
|
1.101
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr6_+_105511615
|
1.091
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr14_-_20098929
|
1.085
|
NM_001001673
NM_001110356
NM_001110357
NM_001110358
NM_001110359
NM_001110360
NM_001110361
|
RNASE9
|
ribonuclease, RNase A family, 9 (non-active)
|
|
chr6_+_28032997
|
1.076
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr12_-_119249938
|
1.074
|
NM_000928
|
PLA2G1B
|
phospholipase A2, group IB (pancreas)
|
|
chr3_+_99733432
|
1.068
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr15_+_70477707
|
1.054
|
NM_001080462
|
TMEM202
|
transmembrane protein 202
|
|
chr12_+_53178834
|
1.051
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr11_+_71221893
|
1.045
|
NM_001002035
|
DEFB108B
|
defensin, beta 108B
|
|
chr6_-_132980901
|
1.040
|
NM_014626
|
TAAR2
|
trace amine associated receptor 2
|
|
chr9_-_21358005
|
1.039
|
NM_006900
|
IFNA1
IFNA13
|
interferon, alpha 1
interferon, alpha 13
|
|
chr9_-_21197141
|
1.038
|
NM_002171
|
IFNA10
|
interferon, alpha 10
|
|
chr6_-_136829705
|
1.036
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr19_+_4104593
|
1.035
|
NM_032607
|
CREB3L3
|
cAMP responsive element binding protein 3-like 3
|
|
chr8_+_18072250
|
1.032
|
NM_001160179
|
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase)
|
|
chr13_-_38462857
|
1.029
|
NM_001144033
NM_145286
|
STOML3
|
stomatin (EPB72)-like 3
|
|
chr19_-_45141473
|
1.025
|
|
LOC440525
PRR13
|
proline rich 13 pseudogene
proline rich 13
|
|
chr10_-_69125932
|
1.017
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr11_-_7906784
|
1.016
|
NM_001004461
|
OR10A6
|
olfactory receptor, family 10, subfamily A, member 6
|
|
chr3_+_46258875
|
1.011
|
NM_001164680
NM_001837
NM_178328
NM_178329
|
CCR3
|
chemokine (C-C motif) receptor 3
|
|
chr2_-_38156811
|
1.004
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chrX_-_108611940
|
0.990
|
NM_001522
|
GUCY2F
|
guanylate cyclase 2F, retinal
|
|
chr15_-_48192804
|
0.978
|
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr3_-_126412882
|
0.978
|
NM_001195483
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr7_+_139750311
|
0.972
|
NM_001008749
|
RAB19
|
RAB19, member RAS oncogene family
|
|
chr5_+_147672181
|
0.968
|
NM_032566
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative)
|
|
chr6_-_44341296
|
0.963
|
NM_004556
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr3_+_131175803
|
0.955
|
NM_007117
|
TRH
|
thyrotropin-releasing hormone
|
|
chr8_-_81155564
|
0.954
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr18_-_68362702
|
0.952
|
NM_182511
|
CBLN2
|
cerebellin 2 precursor
|
|
chr11_-_104332631
|
0.951
|
NM_033306
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr3_-_152541129
|
0.937
|
NM_176876
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12
|
|
chr1_+_149275652
|
0.935
|
NM_001159642
NM_138278
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like
|
|
chr1_-_150353179
|
0.929
|
NM_007113
|
TCHH
|
trichohyalin
|
|
chr9_+_5880801
|
0.926
|
NM_005511
|
MLANA
|
melan-A
|
|
chr6_+_46869052
|
0.926
|
NM_005588
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase)
|
|
chr2_-_219906142
|
0.919
|
NM_001007089
|
RESP18
|
regulated endocrine-specific protein 18 homolog (rat)
|
|
chr7_-_16888121
|
0.911
|
NM_176813
|
AGR3
|
anterior gradient homolog 3 (Xenopus laevis)
|
|
chr9_+_134843918
|
0.903
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr6_+_49575629
|
0.891
|
NM_001010904
|
GLYATL3
|
glycine-N-acyltransferase-like 3
|
|
chr7_-_38279757
|
0.888
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr10_-_45131061
|
0.888
|
NM_001004297
|
OR13A1
|
olfactory receptor, family 13, subfamily A, member 1
|
|
chr10_+_90511142
|
0.879
|
NM_001102469
|
LIPN
|
lipase, family member N
|
|
chr5_+_68823874
|
0.878
|
NM_002538
|
OCLN
|
occludin
|
|
chr2_-_207291364
|
0.875
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr12_-_25597444
|
0.871
|
NM_152590
NM_001145728
NM_001145729
|
IFLTD1
|
intermediate filament tail domain containing 1
|
|
chr9_-_115877104
|
0.864
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr3_+_99699137
|
0.854
|
NM_001004737
|
OR5K2
|
olfactory receptor, family 5, subfamily K, member 2
|
|
chr5_-_177140071
|
0.849
|
NM_173663
|
FAM153A
FAM153C
|
family with sequence similarity 153, member A
family with sequence similarity 153, member C
|
|
chr1_+_205328834
|
0.849
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr11_-_13474101
|
0.844
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr7_+_28305464
|
0.840
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_-_135318420
|
0.833
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr11_-_5756472
|
0.828
|
NM_001001922
|
OR52N5
|
olfactory receptor, family 52, subfamily N, member 5
|
|
chr11_-_8921155
|
0.828
|
NM_020646
|
ASCL3
|
achaete-scute complex homolog 3 (Drosophila)
|
|
chr11_-_18018880
|
0.828
|
NM_004179
|
TPH1
|
tryptophan hydroxylase 1
|
|
chr8_+_12847521
|
0.825
|
NM_020844
|
KIAA1456
|
KIAA1456
|
|
chr12_+_9871343
|
0.804
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr12_-_10850845
|
0.800
|
NM_023918
|
TAS2R8
|
taste receptor, type 2, member 8
|
|
chr10_+_8137356
|
0.800
|
|
GATA3
|
GATA binding protein 3
|
|
chr3_+_12367993
|
0.799
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr11_+_26310253
|
0.798
|
NM_031418
|
ANO3
|
anoctamin 3
|
|
chr17_+_41424665
|
0.796
|
|
MAPT
|
microtubule-associated protein tau
|
|
chr12_-_9777126
|
0.791
|
NM_172004
|
CLECL1
|
C-type lectin-like 1
|
|
chr14_-_98807307
|
0.788
|
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr10_+_63950212
|
0.787
|
NM_199452
|
ZNF365
|
zinc finger protein 365
|
|
chr8_-_120720200
|
0.786
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr4_+_74566325
|
0.786
|
NM_001133
|
AFM
|
afamin
|
|
chr1_-_142559339
|
0.785
|
NM_001123068
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G
|
|
chr4_+_100651183
|
0.782
|
NM_032149
|
C4orf17
|
chromosome 4 open reading frame 17
|
|
chr4_+_71126376
|
0.779
|
NM_152997
|
C4orf7
|
chromosome 4 open reading frame 7
|
|
chr7_-_100630912
|
0.773
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr5_-_147142457
|
0.764
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr2_+_109907623
|
0.764
|
NM_001037866
NM_001123363
NM_032260
|
RGPD6
RGPD5
|
RANBP2-like and GRIP domain containing 6
RANBP2-like and GRIP domain containing 5
|
|
chr9_-_70780574
|
0.760
|
|
|
|
|
chr10_+_90552466
|
0.758
|
NM_001128215
|
LIPM
|
lipase, family member M
|
|
chr5_-_147142368
|
0.757
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr6_-_76838944
|
0.755
|
NM_001563
|
IMPG1
|
interphotoreceptor matrix proteoglycan 1
|
|
chr15_-_48345453
|
0.748
|
NM_002112
|
HDC
|
histidine decarboxylase
|
|
chr15_+_56217686
|
0.748
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr6_-_117853710
|
0.745
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr17_-_30909219
|
0.737
|
NM_001129820
|
SLFN14
|
schlafen family member 14
|
|
chr19_+_61624
|
0.733
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr12_+_52618815
|
0.732
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr12_+_10222823
|
0.725
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr5_+_54434230
|
0.725
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr18_+_59405513
|
0.724
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr12_-_10953427
|
0.721
|
NM_023920
|
TAS2R13
|
taste receptor, type 2, member 13
|
|
chr19_+_6415259
|
0.717
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr1_+_58899
|
0.716
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chr4_+_22303645
|
0.714
|
NM_001128432
NM_020973
|
GBA3
|
glucosidase, beta, acid 3 (cytosolic)
|
|
chr5_+_115415061
|
0.713
|
NM_001195581
|
LOC644100
|
hypothetical LOC644100
|
|
chr4_+_155707089
|
0.706
|
|
FGB
|
fibrinogen beta chain
|
|
chr6_+_132933153
|
0.705
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chr1_+_119712918
|
0.705
|
NM_001005783
NM_016527
|
HAO2
|
hydroxyacid oxidase 2 (long chain)
|
|
chr1_-_167969802
|
0.704
|
NM_000450
|
SELE
|
selectin E
|
|
chr7_-_124217978
|
0.702
|
NM_001024603
|
LOC154872
|
hypothetical protein LOC154872
|
|
chr1_+_198278339
|
0.696
|
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr8_+_23157094
|
0.692
|
NM_152272
|
CHMP7
|
CHMP family, member 7
|
|
chr4_-_157006827
|
0.691
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chr11_-_70350292
|
0.685
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr4_+_40446670
|
0.680
|
NM_024677
|
NSUN7
|
NOP2/Sun domain family, member 7
|
|
chr3_-_121445831
|
0.677
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr6_-_89984214
|
0.676
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr7_+_100169184
|
0.676
|
NM_003386
NM_173059
|
ZAN
|
zonadhesin
|
|
chr4_+_14950657
|
0.674
|
NM_001135170
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr8_+_62363078
|
0.670
|
NM_173519
|
CLVS1
|
clavesin 1
|
|
chr10_+_115514567
|
0.666
|
NM_182601
|
C10orf81
|
chromosome 10 open reading frame 81
|
|
chr8_+_86287161
|
0.665
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr14_-_60818263
|
0.665
|
NM_001017970
|
TMEM30B
|
transmembrane protein 30B
|
|
chr3_-_55489973
|
0.661
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr21_-_26867138
|
0.661
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr12_+_68028397
|
0.658
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr19_+_9984861
|
0.652
|
NM_015725
|
RDH8
|
retinol dehydrogenase 8 (all-trans)
|
|
chr8_+_24207524
|
0.650
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr9_-_111253484
|
0.646
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr9_-_116196505
|
0.646
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr17_-_36577949
|
0.639
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr6_+_32114060
|
0.639
|
NM_000500
NM_001128590
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2
|
|
chr22_-_48866484
|
0.636
|
NM_015166
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr8_+_24297742
|
0.636
|
NM_001145271
NM_001145272
NM_014479
|
ADAMDEC1
|
ADAM-like, decysin 1
|
|
chr10_+_51219558
|
0.634
|
NM_002443
NM_138634
|
MSMB
|
microseminoprotein, beta-
|
|
chr13_-_32678142
|
0.634
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr19_+_20049800
|
0.632
|
NM_007138
|
ZNF90
|
zinc finger protein 90
|
|
chr10_+_106024638
|
0.630
|
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr3_+_99288706
|
0.630
|
NM_054106
|
OR5AC2
|
olfactory receptor, family 5, subfamily AC, member 2
|
|
chr18_+_606671
|
0.629
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr21_+_30895276
|
0.628
|
NM_181620
|
KRTAP22-1
|
keratin associated protein 22-1
|
|
chr1_+_116181927
|
0.628
|
|
NHLH2
|
nescient helix loop helix 2
|
|
chr1_-_244647333
|
0.624
|
NM_022743
|
SMYD3
|
SET and MYND domain containing 3
|
|
chr11_+_122214415
|
0.623
|
NM_019604
|
CRTAM
|
cytotoxic and regulatory T cell molecule
|
|
chr2_-_46988449
|
0.621
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr8_-_37916803
|
0.621
|
NM_152413
|
GOT1L1
|
glutamic-oxaloacetic transaminase 1-like 1
|
|
chrX_-_153903963
|
0.619
|
NM_000132
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr12_-_69434650
|
0.618
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr3_+_131761867
|
0.616
|
NM_001102608
|
COL6A6
|
collagen, type VI, alpha 6
|
|
chr4_+_68995761
|
0.614
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr10_-_123343320
|
0.612
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr1_+_89602023
|
0.611
|
NM_198460
|
GBP6
|
guanylate binding protein family, member 6
|