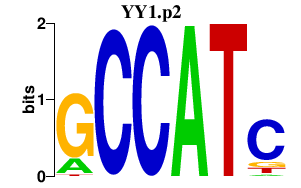
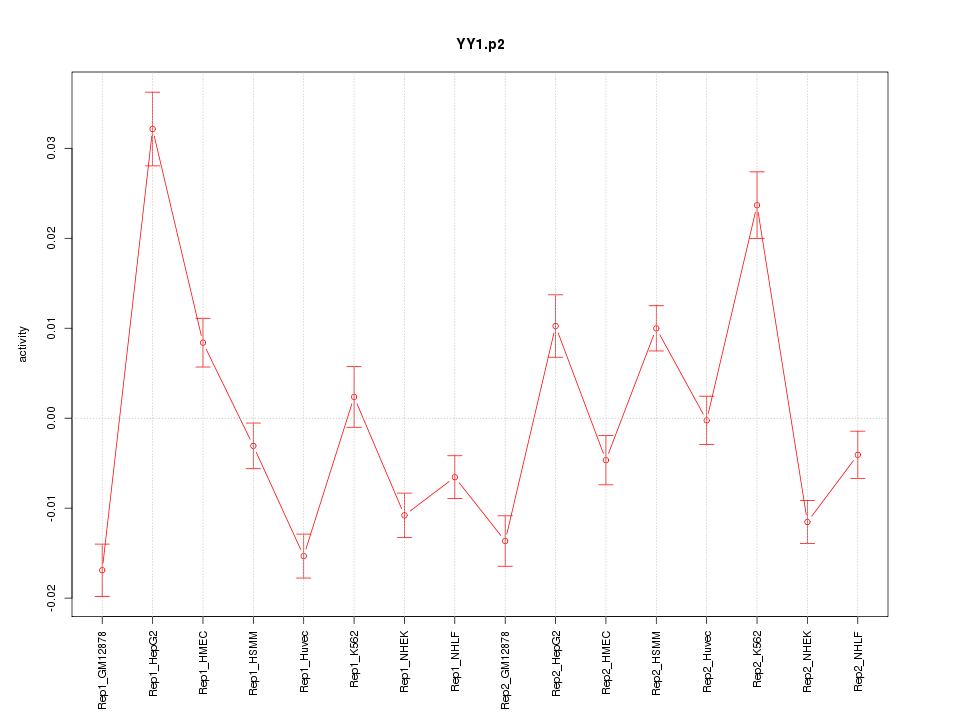
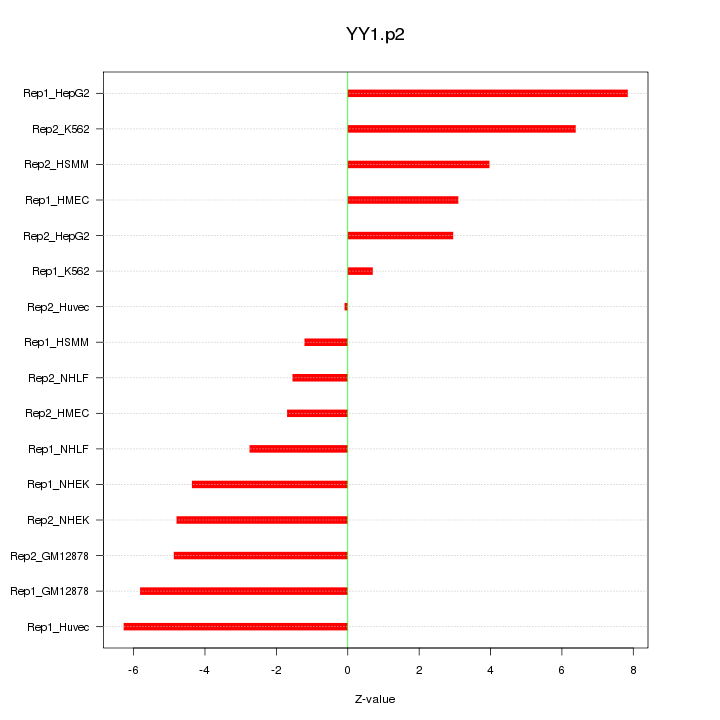
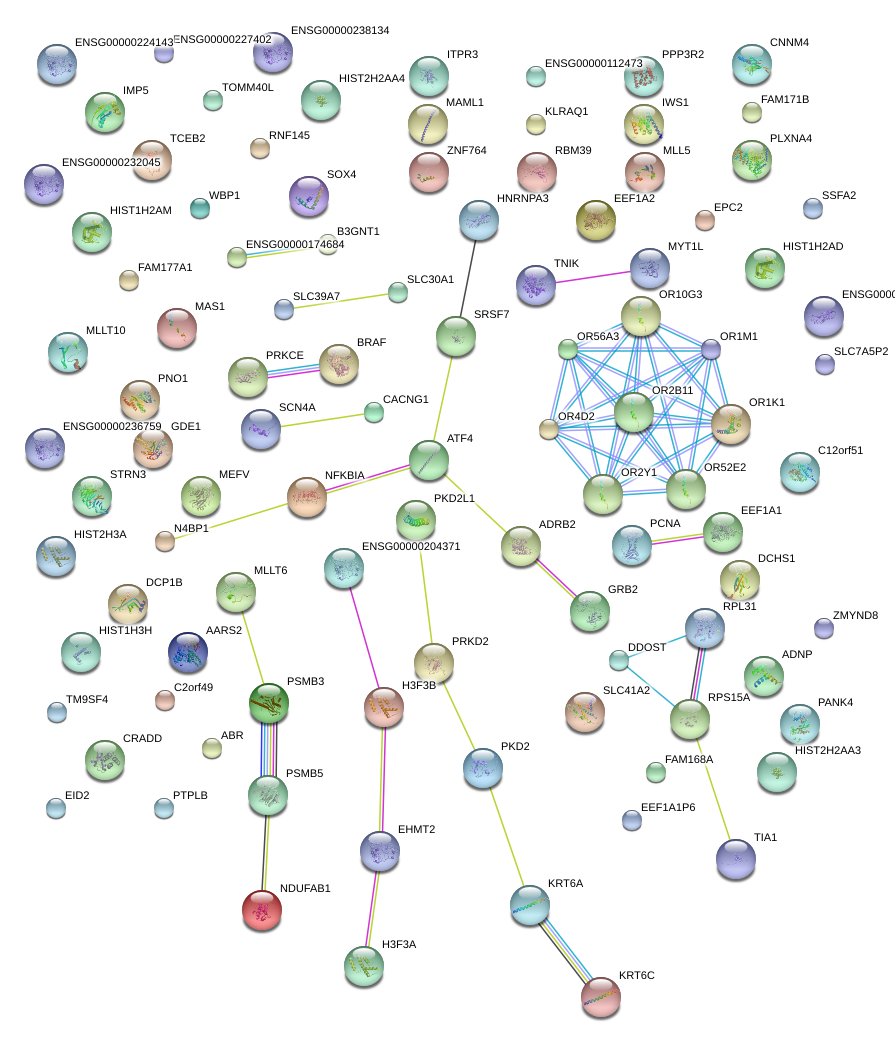
|
chr2_-_1905508
|
6.918
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr16_-_30841938
|
6.099
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr6_+_21701942
|
5.912
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr14_-_22574193
|
5.546
|
NM_001144932
NM_002797
NM_001130725
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr1_-_245681906
|
5.502
|
NM_001004492
|
OR2B11
|
olfactory receptor, family 2, subfamily B, member 11
|
|
chr2_+_149119029
|
5.043
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr17_+_41278035
|
5.004
|
NM_175882
|
IMP5
|
intramembrane protease 5
|
|
chr14_-_22573764
|
3.971
|
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr16_-_21439195
|
3.944
|
|
SLC7A5P2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 pseudogene 2
|
|
chr2_+_48521411
|
3.836
|
NM_001135629
NM_001193475
NM_152994
|
KLRAQ1
|
KLRAQ motif containing 1
|
|
chr20_+_30160952
|
3.779
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr1_-_209818342
|
3.659
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr5_+_148186348
|
3.620
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr20_+_30161173
|
3.496
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr7_-_131843377
|
3.377
|
|
PLXNA4
|
plexin A4
|
|
chr6_+_33276580
|
3.351
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr2_+_96790298
|
3.276
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr2_+_68238490
|
3.274
|
NM_020143
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr11_-_5037432
|
3.202
|
NM_001005164
|
OR52E2
|
olfactory receptor, family 52, subfamily E, member 2
|
|
chr2_-_70329067
|
3.185
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr16_-_3246587
|
3.177
|
NM_000243
NM_001198536
|
MEFV
|
Mediterranean fever
|
|
chr20_-_45418818
|
3.077
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr20_-_48980858
|
3.004
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr6_-_74284522
|
2.999
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr16_-_2767129
|
2.911
|
NM_007108
NM_207013
|
TCEB2
|
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B)
|
|
chr9_+_124602190
|
2.891
|
NM_080859
|
OR1K1
|
olfactory receptor, family 1, subfamily K, member 1
|
|
chr3_-_124786526
|
2.808
|
NM_198402
|
PTPLB
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b
|
|
chr5_-_180099663
|
2.804
|
NM_001001657
|
OR2Y1
|
olfactory receptor, family 2, subfamily Y, member 1
|
|
chr16_-_47201588
|
2.802
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr14_-_21108714
|
2.787
|
NM_001005465
|
OR10G3
|
olfactory receptor, family 10, subfamily G, member 3
|
|
chr17_+_53602013
|
2.745
|
NM_001004707
|
OR4D2
|
olfactory receptor, family 4, subfamily D, member 2
|
|
chr1_-_148080888
|
2.623
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr5_-_158569067
|
2.608
|
|
RNF145
|
ring finger protein 145
|
|
chr2_+_45732416
|
2.607
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr9_-_103397023
|
2.574
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr2_-_128000315
|
2.536
|
NM_017969
|
IWS1
|
IWS1 homolog (S. cerevisiae)
|
|
chr2_+_149118791
|
2.535
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr12_-_111304138
|
2.521
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr17_-_70901303
|
2.508
|
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr22_+_38246509
|
2.480
|
NM_001675
NM_182810
|
ATF4
|
activating transcription factor 4 (tax-responsive enhancer element B67)
|
|
chr3_-_172660545
|
2.476
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr6_-_44388964
|
2.476
|
NM_020745
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr20_-_5048443
|
2.378
|
NM_182649
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr5_-_158567411
|
2.351
|
NM_144726
|
RNF145
|
ring finger protein 145
|
|
chr1_+_159462456
|
2.347
|
NM_032174
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like
|
|
chr17_-_71285992
|
2.342
|
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr17_+_34162525
|
2.334
|
NM_002795
|
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3
|
|
chr14_-_34943574
|
2.311
|
NM_020529
|
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr16_-_30477006
|
2.307
|
NM_001172679
NM_033410
|
ZNF764
|
zinc finger protein 764
|
|
chr11_-_65871592
|
2.294
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr2_+_74539039
|
2.293
|
NM_012477
|
WBP1
|
WW domain binding protein 1
|
|
chr11_+_5925152
|
2.284
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr14_-_30565275
|
2.282
|
NM_001083893
NM_014574
|
STRN3
|
striatin, calmodulin binding protein 3
|
|
chr6_-_31973418
|
2.219
|
|
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2
|
|
chr17_+_34114924
|
2.214
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr2_+_182464910
|
2.213
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr19_-_44722599
|
2.199
|
NM_153232
|
EID2
|
EP300 interacting inhibitor of differentiation 2
|
|
chr4_+_89147821
|
2.198
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr16_-_23515104
|
2.195
|
NM_005003
|
NDUFAB1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa
|
|
chr6_+_33697023
|
2.162
|
NM_002224
|
ITPR3
|
inositol 1,4,5-triphosphate receptor, type 3
|
|
chr5_+_179092405
|
2.154
|
NM_014757
|
MAML1
|
mastermind-like 1 (Drosophila)
|
|
chr2_+_100985122
|
2.141
|
NM_000993
NM_001098577
NM_001099693
|
RPL31
|
ribosomal protein L31
|
|
chr12_-_103846186
|
2.137
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr2_+_177785667
|
2.127
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr11_-_6600385
|
2.112
|
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr16_-_19440786
|
2.108
|
NM_016641
|
GDE1
|
glycerophosphodiester phosphodiesterase 1
|
|
chr17_-_975809
|
2.101
|
|
ABR
|
active BCR-related gene
|
|
chr6_+_160247962
|
2.084
|
NM_002377
|
MAS1
|
MAS1 oncogene
|
|
chr1_-_2447848
|
2.082
|
NM_018216
|
PANK4
|
pantothenate kinase 4
|
|
chr2_-_38831901
|
2.066
|
NM_001031684
NM_001195446
|
SRSF7
|
serine/arginine-rich splicing factor 7
|
|
chr20_-_33793507
|
1.993
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr2_+_105320440
|
1.992
|
NM_024093
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr17_-_59403932
|
1.991
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chr11_-_72986738
|
1.982
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr2_+_187266899
|
1.978
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr19_-_52307642
|
1.974
|
|
|
|
|
chr16_-_18709125
|
1.959
|
NM_001019
NM_001030009
|
RPS15A
|
ribosomal protein S15a
|
|
chr12_-_51173238
|
1.947
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr12_+_92595281
|
1.947
|
NM_003805
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain
|
|
chr7_-_140270749
|
1.912
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr11_+_76491532
|
1.910
|
NM_006189
|
OMP
|
olfactory marker protein
|
|
chr17_+_62471167
|
1.909
|
NM_000727
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1
|
|
chr7_+_104441730
|
1.904
|
NM_018682
NM_182931
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr14_+_34585098
|
1.902
|
NM_173607
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr19_+_9064854
|
1.897
|
NM_001004456
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1
|
|
chr6_-_31973434
|
1.880
|
NM_006709
NM_025256
|
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2
|
|
chr12_-_1983845
|
1.876
|
NM_152640
|
DCP1B
|
DCP1 decapping enzyme homolog B (S. cerevisiae)
|
|
chr17_-_45619038
|
1.867
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr17_-_40122542
|
1.861
|
NM_001099225
NM_144609
|
CCDC43
|
coiled-coil domain containing 43
|
|
chr8_-_145097031
|
1.853
|
NM_201380
|
PLEC
|
plectin
|
|
chr1_-_12830728
|
1.835
|
NM_001146181
|
HNRNPCL1
LOC649330
|
heterogeneous nuclear ribonucleoprotein C-like 1
heterogeneous nuclear ribonucleoprotein C-like
|
|
chr2_+_183697327
|
1.826
|
NM_138285
|
NUP35
|
nucleoporin 35kDa
|
|
chr11_-_68505030
|
1.810
|
NM_198923
|
MRGPRD
|
MAS-related GPR, member D
|
|
chr6_-_116681863
|
1.808
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr9_-_110815542
|
1.806
|
NM_003798
|
CTNNAL1
|
catenin (cadherin-associated protein), alpha-like 1
|
|
chr12_-_120725210
|
1.804
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr7_+_148523486
|
1.795
|
NM_003575
|
ZNF282
|
zinc finger protein 282
|
|
chr14_-_34001164
|
1.793
|
NM_138288
|
C14orf147
|
chromosome 14 open reading frame 147
|
|
chr19_-_60587414
|
1.792
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr17_-_37275142
|
1.792
|
NM_018143
|
KLHL11
|
kelch-like 11 (Drosophila)
|
|
chr16_-_29532538
|
1.789
|
|
|
|
|
chr11_+_4522993
|
1.786
|
NM_001004137
|
OR52M1
|
olfactory receptor, family 52, subfamily M, member 1
|
|
chr16_-_86460529
|
1.775
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5
|
|
chr6_-_27222556
|
1.770
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr11_+_4466684
|
1.770
|
NM_001005171
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1
|
|
chr2_-_207738858
|
1.758
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr6_+_26153590
|
1.752
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr14_+_22860547
|
1.729
|
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr16_-_30529129
|
1.726
|
NM_138447
|
ZNF689
|
zinc finger protein 689
|
|
chr20_+_10363949
|
1.723
|
NM_001009608
|
C20orf94
|
chromosome 20 open reading frame 94
|
|
chr6_-_152664906
|
1.712
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr1_+_178190493
|
1.710
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr3_-_102888090
|
1.705
|
NM_000986
|
RPL24
|
ribosomal protein L24
|
|
chr17_-_53784494
|
1.700
|
NM_003168
|
SUPT4H1
|
suppressor of Ty 4 homolog 1 (S. cerevisiae)
|
|
chr6_-_88911708
|
1.695
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr2_-_240618518
|
1.690
|
NM_001005853
|
OR6B2
|
olfactory receptor, family 6, subfamily B, member 2
|
|
chr14_+_22845851
|
1.688
|
NM_004050
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 read-through transcript
BCL2-like 2
|
|
chr20_-_472335
|
1.685
|
NM_001895
NM_177559
NM_177560
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide
|
|
chr9_-_114858788
|
1.680
|
NM_003408
|
ZFP37
|
zinc finger protein 37 homolog (mouse)
|
|
chr6_+_27208763
|
1.678
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr4_-_23500706
|
1.674
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr6_+_30142910
|
1.652
|
NM_021959
|
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11
|
|
chr11_-_65962836
|
1.651
|
NM_016050
NM_170738
NM_170739
|
MRPL11
|
mitochondrial ribosomal protein L11
|
|
chr13_+_48582389
|
1.648
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr11_+_5862076
|
1.637
|
NM_001005165
|
OR52E4
|
olfactory receptor, family 52, subfamily E, member 4
|
|
chr20_-_33793286
|
1.635
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr1_+_144218960
|
1.633
|
NM_005105
|
RBM8A
|
RNA binding motif protein 8A
|
|
chr10_-_32707549
|
1.623
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr17_-_57497380
|
1.617
|
NM_005121
|
MED13
|
mediator complex subunit 13
|
|
chr11_-_66976775
|
1.614
|
NM_206997
|
GPR152
|
G protein-coupled receptor 152
|
|
chr15_-_39195701
|
1.612
|
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr14_+_30161285
|
1.610
|
|
SCFD1
|
sec1 family domain containing 1
|
|
chr6_-_112301123
|
1.598
|
NM_002037
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr6_-_26140266
|
1.585
|
NM_003537
|
HIST1H3B
|
histone cluster 1, H3b
|
|
chr11_+_4885138
|
1.578
|
NM_001004749
|
OR51A7
|
olfactory receptor, family 51, subfamily A, member 7
|
|
chr1_+_45822246
|
1.578
|
NM_001195193
NM_002482
NM_152298
|
NASP
|
nuclear autoantigenic sperm protein (histone-binding)
|
|
chr2_-_178125680
|
1.575
|
NM_152517
|
TTC30B
|
tetratricopeptide repeat domain 30B
|
|
chr17_+_42023353
|
1.573
|
NM_006178
|
NSF
|
N-ethylmaleimide-sensitive factor
|
|
chr2_+_176724358
|
1.573
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr2_+_46560207
|
1.569
|
NM_001145051
|
LOC388946
|
transmembrane protein ENSP00000343375
|
|
chr4_+_9094611
|
1.568
|
NM_001040071
|
LOC650293
|
seven transmembrane helix receptor
|
|
chr6_+_30799130
|
1.567
|
|
TUBB
|
tubulin, beta
|
|
chr1_+_42896634
|
1.565
|
NM_006347
|
PPIH
|
peptidylprolyl isomerase H (cyclophilin H)
|
|
chr2_+_105320247
|
1.564
|
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr15_-_81667169
|
1.561
|
NM_016073
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr11_+_113815317
|
1.556
|
NM_015523
|
REXO2
|
REX2, RNA exonuclease 2 homolog (S. cerevisiae)
|
|
chr5_-_180603335
|
1.555
|
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1
|
|
chr12_+_100615530
|
1.540
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr17_+_40494167
|
1.535
|
|
NMT1
|
N-myristoyltransferase 1
|
|
chr2_+_241279934
|
1.531
|
NM_198998
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr2_+_177965734
|
1.526
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr12_+_55148567
|
1.524
|
NM_207344
|
SPRYD4
|
SPRY domain containing 4
|
|
chr4_+_39376058
|
1.522
|
NM_001111112
NM_001111113
NM_005339
|
UBE2K
|
ubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast)
|
|
chr6_+_133177403
|
1.518
|
|
RPS12
|
ribosomal protein S12
|
|
chr12_+_74160779
|
1.513
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr1_+_39229474
|
1.503
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr14_+_22305570
|
1.503
|
NM_005015
|
OXA1L
|
oxidase (cytochrome c) assembly 1-like
|
|
chr14_+_30161267
|
1.500
|
NM_016106
NM_182835
|
SCFD1
|
sec1 family domain containing 1
|
|
chr15_-_49817090
|
1.499
|
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2
|
|
chr16_+_21926939
|
1.492
|
NM_001164579
|
C16orf52
|
chromosome 16 open reading frame 52
|
|
chr19_-_57223329
|
1.489
|
NM_025040
|
ZNF614
|
zinc finger protein 614
|
|
chr17_-_72244231
|
1.486
|
|
|
|
|
chr19_+_15699833
|
1.485
|
NM_013939
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2
|
|
chr17_+_39990124
|
1.484
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr8_-_10550026
|
1.473
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr11_+_74788200
|
1.467
|
NM_001005
|
RPS3
|
ribosomal protein S3
|
|
chr4_-_140696740
|
1.466
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr16_-_69114931
|
1.462
|
NM_001195139
NM_015386
|
COG4
|
component of oligomeric golgi complex 4
|
|
chr6_+_27883877
|
1.461
|
NM_003509
|
HIST1H2AI
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H3f
|
|
chr15_-_39195621
|
1.457
|
NM_017553
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr14_-_38709161
|
1.448
|
NM_001079537
NM_177452
|
TRAPPC6B
|
trafficking protein particle complex 6B
|
|
chr20_-_33343617
|
1.447
|
NM_178468
|
FAM83C
|
family with sequence similarity 83, member C
|
|
chr1_-_246857051
|
1.434
|
NM_001001964
|
OR2T11
|
olfactory receptor, family 2, subfamily T, member 11
|
|
chr16_+_70437394
|
1.431
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr11_+_66952462
|
1.422
|
NM_003952
|
RPS6KB2
|
ribosomal protein S6 kinase, 70kDa, polypeptide 2
|
|
chr7_+_128651976
|
1.418
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr6_-_26324850
|
1.416
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr16_-_23068010
|
1.410
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr6_+_30420878
|
1.410
|
NM_024839
|
RPP21
|
ribonuclease P/MRP 21kDa subunit
|
|
chr18_+_11841388
|
1.405
|
NM_020412
|
CHMP1B
|
chromatin modifying protein 1B
|
|
chr6_-_144458326
|
1.404
|
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa
|
|
chr2_+_172087102
|
1.398
|
NM_001127383
NM_024843
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr7_+_142270634
|
1.393
|
|
EPHB6
|
EPH receptor B6
|
|
chr2_+_62787534
|
1.387
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr6_+_133177399
|
1.384
|
NM_001016
|
SNORA33
RPS12
|
small nucleolar RNA, H/ACA box 33
ribosomal protein S12
|
|
chr20_+_1047216
|
1.379
|
NM_006814
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31)
|
|
chr17_-_36915329
|
1.376
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr2_-_55698217
|
1.375
|
NM_001122964
NM_020463
|
SMEK2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium)
|
|
chr17_+_72065440
|
1.372
|
|
LOC100507246
|
hypothetical LOC100507246
|
|
chr11_+_6823458
|
1.370
|
NM_178168
|
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5
|
|
chr4_-_187713512
|
1.369
|
NM_005958
|
MTNR1A
|
melatonin receptor 1A
|
|
chr17_+_45778381
|
1.369
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr10_-_69762654
|
1.366
|
NM_001033083
NM_022129
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr7_+_115953582
|
1.363
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr17_-_53847937
|
1.363
|
|
RNF43
|
ring finger protein 43
|
|
chr10_+_75211810
|
1.362
|
NM_203298
|
CHCHD1
|
coiled-coil-helix-coiled-coil-helix domain containing 1
|
|
chr11_+_4427100
|
1.356
|
NM_001005172
|
OR52K2
|
olfactory receptor, family 52, subfamily K, member 2
|
|
chr5_+_52131686
|
1.349
|
|
PELO
|
pelota homolog (Drosophila)
|
|
chr2_-_60634271
|
1.347
|
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr2_+_172087002
|
1.346
|
|
CYBRD1
|
cytochrome b reductase 1
|