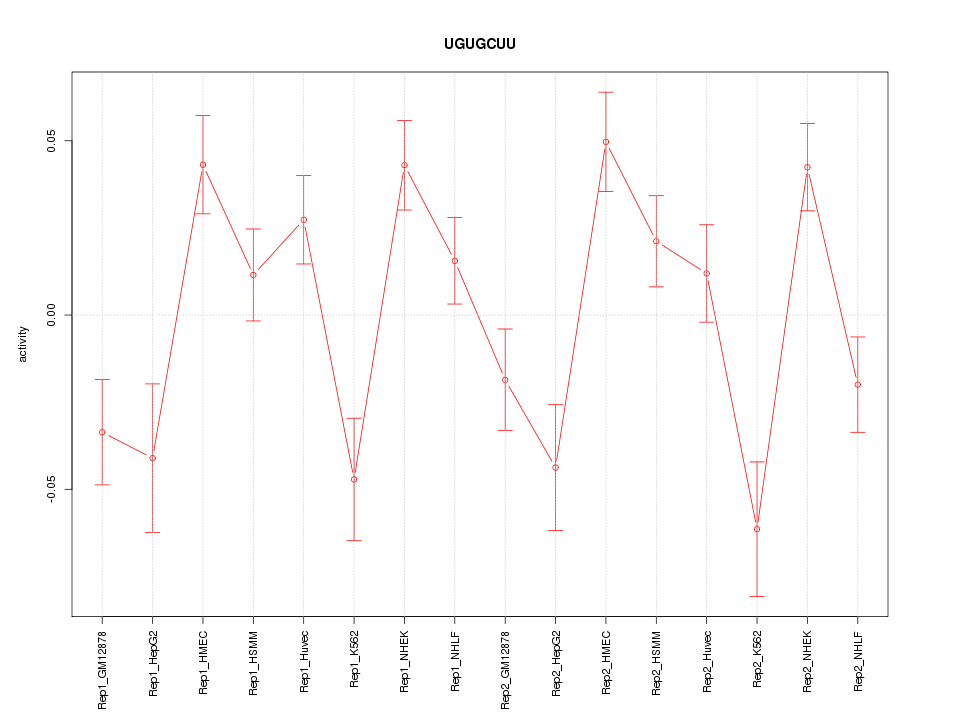
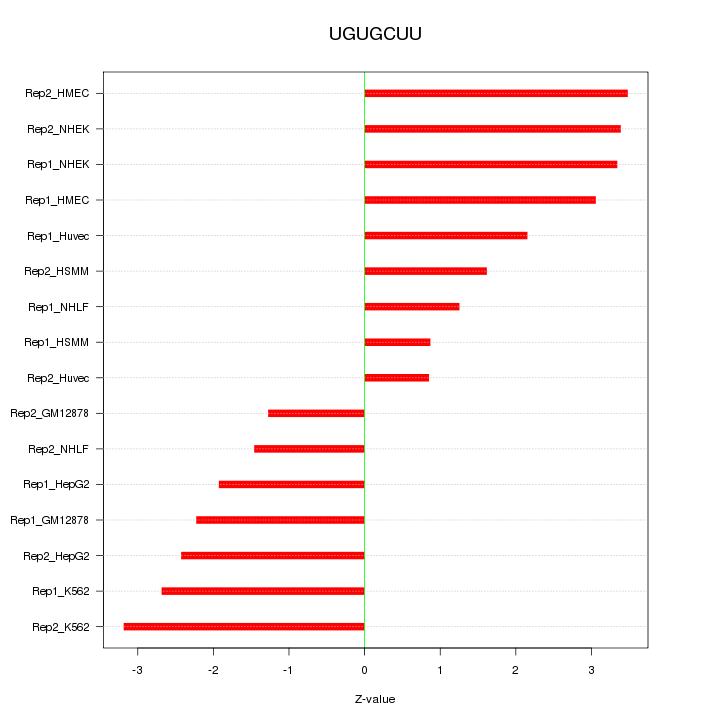
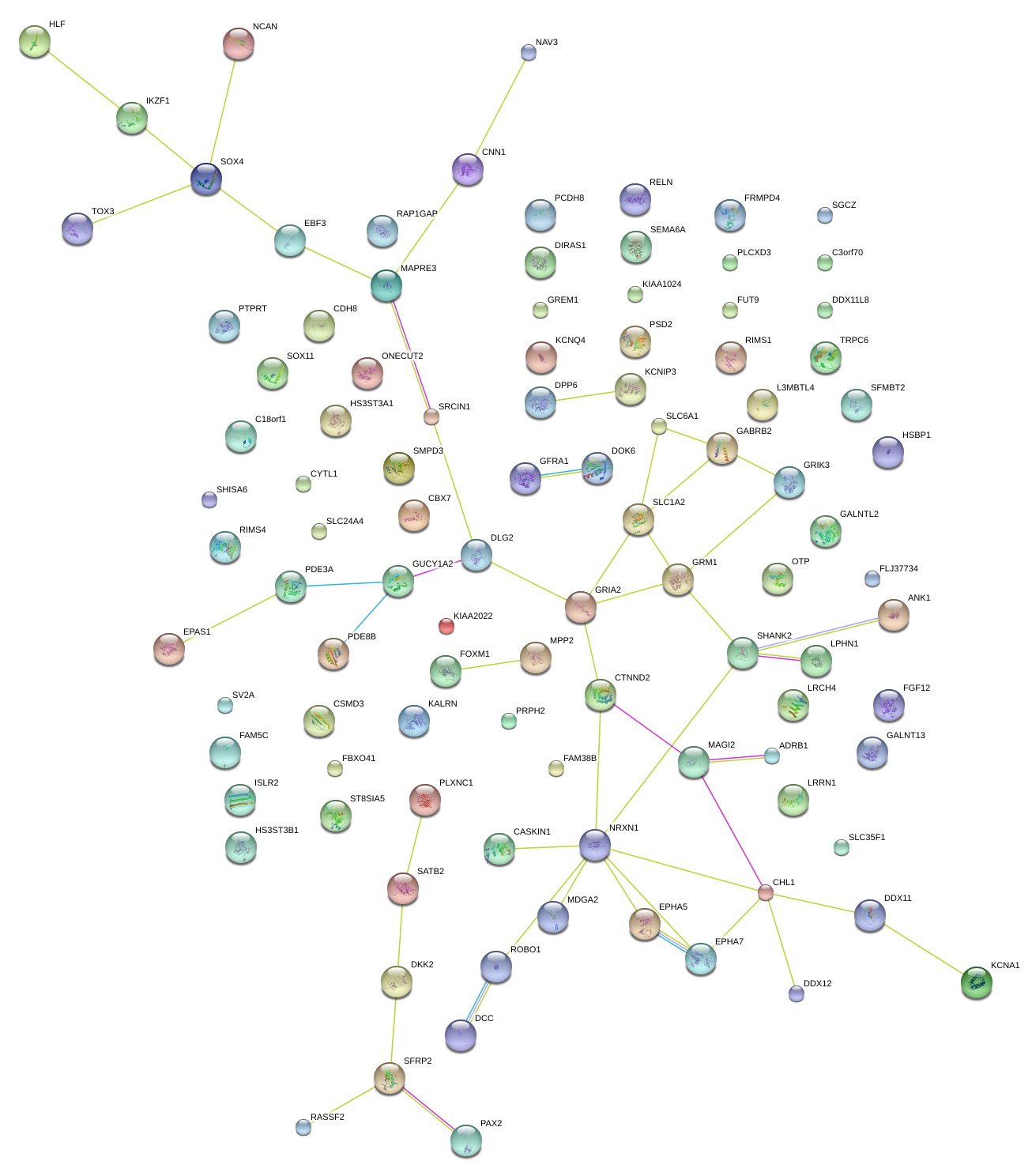
|
chr18_+_53253775
|
4.751
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr2_+_5750229
|
4.724
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr14_-_47213906
|
4.511
|
NM_001113498
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr18_-_6404900
|
4.188
|
NM_173464
|
L3MBTL4
|
l(3)mbt-like 4 (Drosophila)
|
|
chr10_-_7493418
|
4.123
|
NM_001029880
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr17_+_11085464
|
4.036
|
NM_001173461
NM_001173462
NM_207386
|
SHISA6
|
shisa homolog 6 (Xenopus laevis)
|
|
chr16_-_60627536
|
4.013
|
NM_001796
|
CDH8
|
cadherin 8, type 2
|
|
chr4_-_66218247
|
3.942
|
NM_004439
NM_182472
|
EPHA5
|
EPH receptor A5
|
|
chr18_-_42590989
|
3.850
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr10_-_7491204
|
3.800
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr12_+_20413445
|
3.723
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr10_-_118021532
|
3.667
|
NM_145793
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr3_-_193609531
|
3.458
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr10_+_115793795
|
3.453
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr14_+_91859904
|
3.361
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr18_+_65219114
|
3.350
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chr11_-_35397675
|
3.347
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr18_-_11138760
|
3.341
|
NM_022068
|
FAM38B
|
family with sequence similarity 38, member B
|
|
chr11_-_100959868
|
3.202
|
NM_004621
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6
|
|
chr12_+_76749199
|
3.186
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr6_-_94185780
|
3.179
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr12_+_4889309
|
3.145
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr10_-_118022965
|
3.132
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr4_+_158361185
|
3.109
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr7_-_103417198
|
3.078
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr2_+_154436670
|
3.048
|
NM_052917
|
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13)
|
|
chr5_-_160907702
|
3.037
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr1_-_21868380
|
3.001
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr18_+_13208777
|
2.988
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr6_+_72979234
|
2.900
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr2_-_200044233
|
2.887
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr10_-_131652007
|
2.870
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr3_+_125786195
|
2.863
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr4_-_108176901
|
2.854
|
NM_014421
|
DKK2
|
dickkopf homolog 2 (Xenopus laevis)
|
|
chr7_+_153380673
|
2.851
|
NM_130797
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr14_+_91859289
|
2.841
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr3_+_213535
|
2.813
|
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr19_-_2672337
|
2.800
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr3_-_79899748
|
2.788
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr3_+_11009419
|
2.767
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr7_+_50314801
|
2.725
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr5_-_41546384
|
2.711
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr11_-_35398077
|
2.693
|
NM_001195728
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr15_+_72208751
|
2.656
|
NM_001130136
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr2_+_95326753
|
2.646
|
NM_013434
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin
|
|
chr17_+_50697319
|
2.635
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr10_-_118022707
|
2.596
|
NM_001145453
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr8_-_114518194
|
2.589
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr5_-_76970264
|
2.580
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr5_-_11956964
|
2.525
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr6_+_72653369
|
2.515
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr11_-_70185559
|
2.496
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr1_-_21850874
|
2.483
|
NM_001145658
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr1_-_148155986
|
2.478
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chrX_-_74062005
|
2.474
|
NM_001008537
|
KIAA2022
|
KIAA2022
|
|
chr15_+_72209795
|
2.453
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr6_+_118335381
|
2.448
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr18_+_13601664
|
2.428
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr2_-_73350103
|
2.425
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr1_-_37272295
|
2.389
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr1_-_188713324
|
2.383
|
NM_199051
|
FAM5C
|
family with sequence similarity 5, member C
|
|
chr2_-_200038055
|
2.372
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr16_-_67039874
|
2.336
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr10_-_128884411
|
2.331
|
NM_001039762
|
FAM196A
|
family with sequence similarity 196, member A
|
|
chr15_+_30797466
|
2.324
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr3_+_3816079
|
2.322
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr11_-_106394039
|
2.302
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr13_-_52320774
|
2.283
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr16_-_51138306
|
2.265
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr8_-_41873401
|
2.245
|
NM_001142446
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr7_-_78920806
|
2.232
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr8_-_15140162
|
2.231
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr10_+_102495327
|
2.231
|
NM_000278
NM_003987
NM_003988
NM_003989
NM_003990
|
PAX2
|
paired box 2
|
|
chr12_+_93066370
|
2.226
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr20_-_4752067
|
2.217
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr11_-_84312085
|
2.189
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr19_-_14177980
|
2.183
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr5_-_115938305
|
2.175
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr20_-_42872295
|
2.157
|
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr2_-_50428395
|
2.149
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr19_+_11510578
|
2.097
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr16_-_2186465
|
2.096
|
NM_020764
|
CASKIN1
|
CASK interacting protein 1
|
|
chr17_-_34015631
|
2.079
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr22_-_37878394
|
2.063
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr20_-_41251878
|
2.061
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chrX_+_12066505
|
2.046
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr19_+_19183763
|
2.042
|
NM_004386
|
NCAN
|
neurocan
|
|
chr6_+_72983183
|
2.004
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr15_+_72209642
|
1.991
|
NM_001130138
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr6_+_72982865
|
1.969
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_41022066
|
1.966
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr15_+_72209039
|
1.964
|
NM_001130137
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr5_+_76542461
|
1.957
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr17_+_14145051
|
1.951
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr11_-_83071084
|
1.932
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr6_+_146390474
|
1.928
|
NM_000838
NM_001114329
|
GRM1
|
glutamate receptor, metabotropic 1
|
|
chr6_+_96570565
|
1.918
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr4_-_154929657
|
1.905
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|
|
chr18_+_48120539
|
1.891
|
NM_005215
|
DCC
|
deleted in colorectal carcinoma
|
|
chr15_+_77511912
|
1.890
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr5_+_139155569
|
1.884
|
NM_032289
|
PSD2
|
pleckstrin and Sec7 domain containing 2
|
|
chr3_-_186353421
|
1.882
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr17_-_43862480
|
1.871
|
NM_001075099
NM_003726
|
SKAP1
|
src kinase associated phosphoprotein 1
|
|
chrX_-_64171317
|
1.850
|
NM_001178032
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr19_+_38804694
|
1.837
|
NM_001127895
NM_001127896
|
CHST8
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8
|
|
chr2_-_51113040
|
1.833
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr4_+_2031036
|
1.831
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr2_+_176689737
|
1.808
|
NM_002148
|
HOXD10
|
homeobox D10
|
|
chr7_+_153215316
|
1.807
|
NM_001039350
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr3_+_77171938
|
1.774
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr2_-_213111525
|
1.774
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr11_-_45264193
|
1.767
|
NM_020826
|
SYT13
|
synaptotagmin XIII
|
|
chr11_-_8147111
|
1.767
|
NM_001135109
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr4_-_10068055
|
1.763
|
NM_053042
|
ZNF518B
|
zinc finger protein 518B
|
|
chrX_+_51091822
|
1.758
|
NM_153183
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10
|
|
chr7_-_36991189
|
1.755
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr14_+_91858677
|
1.749
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr11_+_104986009
|
1.745
|
NM_000829
NM_001077243
NM_001077244
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|
|
chr11_+_108798047
|
1.744
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr2_-_144994349
|
1.744
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr21_+_18539016
|
1.743
|
NM_024944
|
CHODL
|
chondrolectin
|
|
chr15_-_71448657
|
1.717
|
NM_005477
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr16_-_51139214
|
1.680
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr8_-_93177057
|
1.658
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_-_16650697
|
1.643
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr20_-_604822
|
1.636
|
NM_033129
|
SCRT2
|
scratch homolog 2, zinc finger protein (Drosophila)
|
|
chr13_+_34414390
|
1.627
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr7_-_71440143
|
1.620
|
NM_001017440
|
CALN1
|
calneuron 1
|
|
chr13_+_77170470
|
1.605
|
NM_001040153
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr11_-_77730573
|
1.592
|
NM_012296
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr9_-_90983501
|
1.589
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr6_+_69401660
|
1.570
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr13_+_77213295
|
1.563
|
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chrX_-_83329562
|
1.550
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr18_-_33399997
|
1.541
|
NM_001025087
NM_001025088
NM_001025089
NM_020180
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr18_-_24011349
|
1.540
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr1_-_199259447
|
1.520
|
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr11_+_65815940
|
1.513
|
NM_153266
|
TMEM151A
|
transmembrane protein 151A
|
|
chr11_+_8059267
|
1.512
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr10_+_118990573
|
1.510
|
NM_003054
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine), member 2
|
|
chr3_+_49566901
|
1.508
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr19_-_60646041
|
1.502
|
NM_001145176
|
SHISA7
|
shisa homolog 7 (Xenopus laevis)
|
|
chr22_+_24290738
|
1.498
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr1_+_110494654
|
1.462
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr1_+_145479805
|
1.458
|
NM_004326
|
BCL9
|
B-cell CLL/lymphoma 9
|
|
chr9_+_137733851
|
1.444
|
NM_020822
|
KCNT1
|
potassium channel, subfamily T, member 1
|
|
chr1_+_99502435
|
1.434
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr8_-_70909754
|
1.433
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr2_+_190723216
|
1.426
|
NM_001042520
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr1_-_153109349
|
1.424
|
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr1_+_229365296
|
1.407
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr6_+_73388291
|
1.398
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr10_+_52504239
|
1.394
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr9_+_111442888
|
1.384
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr8_-_145530639
|
1.376
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr3_-_193928038
|
1.375
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr1_-_119333701
|
1.369
|
NM_152380
|
TBX15
|
T-box 15
|
|
chr11_+_91724909
|
1.369
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr11_+_74629597
|
1.357
|
NM_001195528
|
LOC441617
|
hypothetical LOC441617
|
|
chr17_+_32368611
|
1.353
|
NM_005568
|
LHX1
|
LIM homeobox 1
|
|
chr12_-_23993830
|
1.340
|
NM_006940
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr4_-_164473107
|
1.334
|
NM_000909
|
NPY1R
|
neuropeptide Y receptor Y1
|
|
chr5_-_35266550
|
1.303
|
NM_000949
|
PRLR
|
prolactin receptor
|
|
chr7_+_97199206
|
1.300
|
NM_003182
NM_013996
NM_013997
NM_013998
|
TAC1
|
tachykinin, precursor 1
|
|
chr15_-_24570010
|
1.300
|
NM_021912
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr5_-_158459276
|
1.294
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr16_+_57056265
|
1.283
|
NM_022910
|
NDRG4
|
NDRG family member 4
|
|
chr8_-_93176618
|
1.245
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_53055078
|
1.244
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr19_-_17660005
|
1.243
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr14_-_73296643
|
1.237
|
NM_194278
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr8_-_93184629
|
1.227
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_152663748
|
1.227
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr2_+_120820140
|
1.224
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr13_-_99422163
|
1.207
|
NM_033132
|
ZIC5
|
Zic family member 5 (odd-paired homolog, Drosophila)
|
|
chr2_-_200030944
|
1.207
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr6_+_101953581
|
1.200
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr14_-_56346849
|
1.197
|
NM_021728
|
OTX2
|
orthodenticle homeobox 2
|
|
chr13_-_35603513
|
1.188
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr12_+_70952729
|
1.182
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr12_-_16652202
|
1.173
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_-_24946495
|
1.171
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr11_+_22316242
|
1.169
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr10_+_52420936
|
1.166
|
NM_001098512
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr5_-_88215034
|
1.155
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_+_231437745
|
1.153
|
NM_001012514
NM_001012516
NM_030926
|
ITM2C
|
integral membrane protein 2C
|
|
chr3_+_23961739
|
1.148
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr8_-_93181091
|
1.104
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr15_-_24569309
|
1.103
|
NM_000814
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr11_-_31796061
|
1.103
|
NM_001127612
|
PAX6
|
paired box 6
|
|
chr9_-_92444886
|
1.102
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr1_-_177378801
|
1.102
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chrX_+_56606686
|
1.098
|
NM_013444
|
UBQLN2
|
ubiquilin 2
|
|
chr7_-_11838260
|
1.087
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr11_-_16380967
|
1.084
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr13_+_95541093
|
1.080
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr12_-_69600780
|
1.070
|
NM_002849
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr7_+_140420500
|
1.067
|
NM_001195278
|
LOC100131199
|
transmembrane protein 178-like
|
|
chr3_+_85090721
|
1.060
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr19_-_45888259
|
1.042
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|