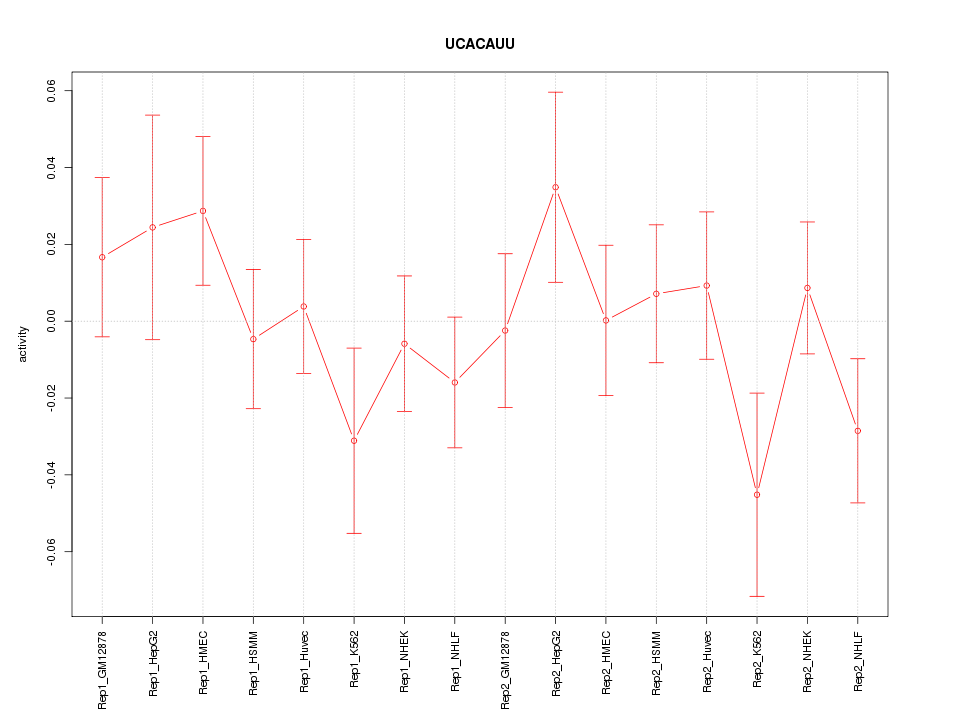
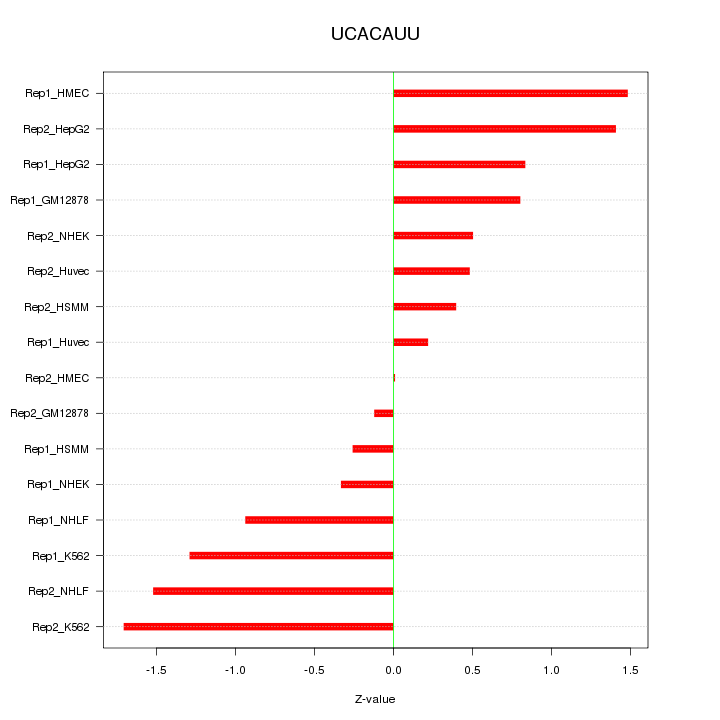
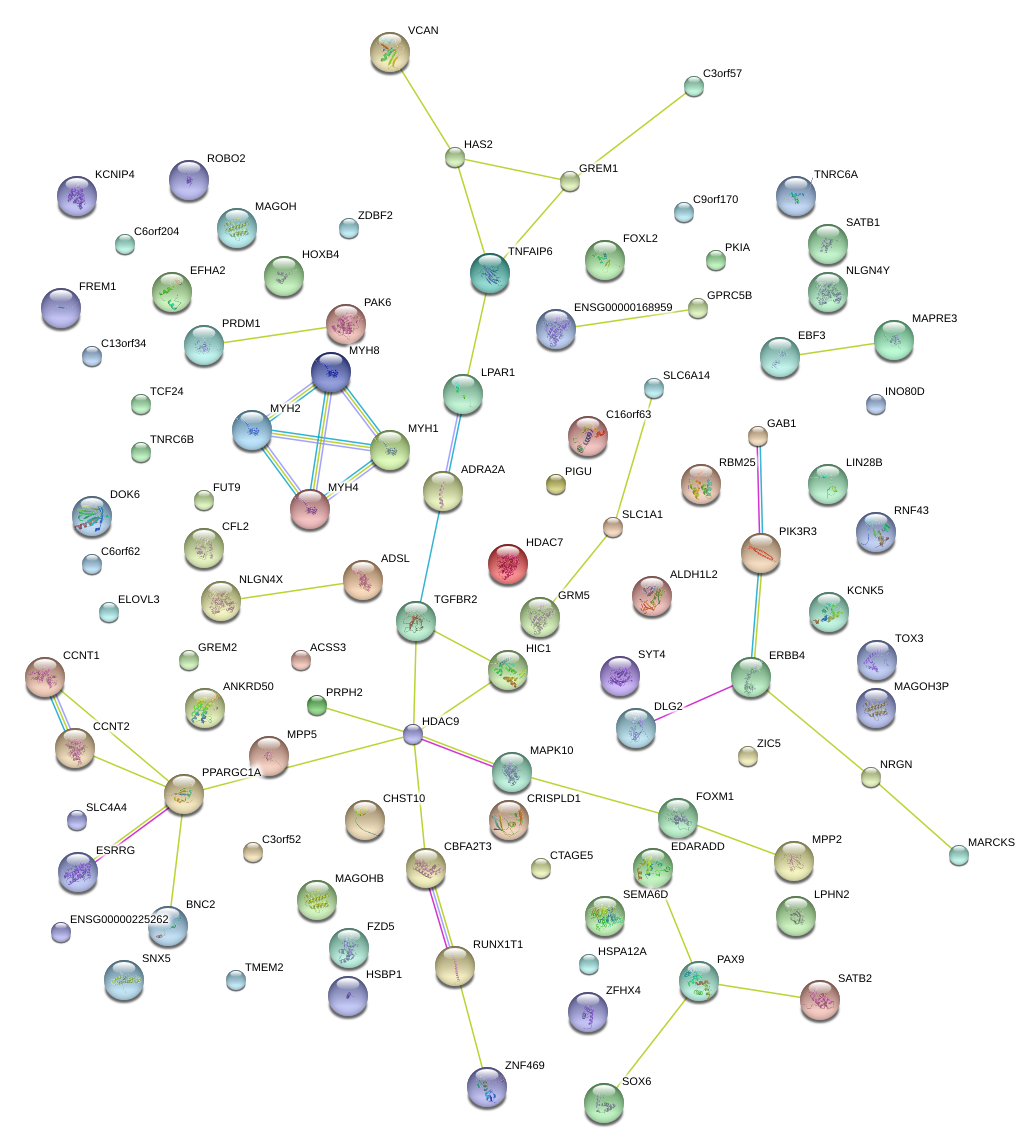
|
chr4_+_72271218
|
1.636
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr2_-_213111525
|
1.603
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr16_-_87570716
|
1.573
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr15_+_30797466
|
1.466
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr2_-_200038055
|
1.325
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr13_-_99422163
|
1.310
|
NM_033132
|
ZIC5
|
Zic family member 5 (odd-paired homolog, Drosophila)
|
|
chr8_-_122722674
|
1.223
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr6_+_96570565
|
1.223
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr2_-_200044233
|
1.208
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr3_+_77171938
|
1.152
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr3_+_30622905
|
1.081
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr22_+_38903825
|
1.012
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr9_-_73573121
|
0.992
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr8_+_76059262
|
0.896
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chrX_-_6156655
|
0.889
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr2_-_100400481
|
0.875
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr11_-_88436463
|
0.865
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr9_-_112840064
|
0.852
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr9_+_4480193
|
0.848
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr18_-_39111243
|
0.753
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chrX_-_6155887
|
0.746
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr1_-_238842071
|
0.709
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chrX_+_115481774
|
0.702
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr8_-_93176618
|
0.690
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_162572441
|
0.674
|
NM_001040100
|
C3orf57
|
chromosome 3 open reading frame 57
|
|
chr6_+_114285209
|
0.620
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr1_+_82038669
|
0.619
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr12_-_104002348
|
0.618
|
NM_001034173
|
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2
|
|
chr14_+_38806028
|
0.607
|
NM_005930
NM_203355
|
CTAGE5
|
CTAGE family, member 5
|
|
chr14_+_36196523
|
0.594
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr2_-_206659140
|
0.589
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr10_-_131652007
|
0.574
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr2_-_200030944
|
0.573
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr16_-_51139214
|
0.572
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr10_-_118491971
|
0.567
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr9_+_88953378
|
0.565
|
NM_001001709
|
C9orf170
|
chromosome 9 open reading frame 170
|
|
chr8_-_93184629
|
0.553
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_-_10313600
|
0.546
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr16_-_19803606
|
0.539
|
NM_016235
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr12_+_79995927
|
0.534
|
NM_024560
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3
|
|
chr4_-_23500706
|
0.534
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr9_-_16860719
|
0.522
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr12_-_10657397
|
0.510
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr2_+_151922350
|
0.508
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr15_+_38319326
|
0.506
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr1_-_46370837
|
0.504
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr4_+_72423633
|
0.504
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_-_125853166
|
0.498
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr4_-_20914626
|
0.496
|
NM_147183
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr14_-_34253473
|
0.495
|
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr15_+_45797977
|
0.492
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr17_-_53849929
|
0.472
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr1_+_234624266
|
0.471
|
NM_145861
|
EDARADD
|
EDAR-associated death domain
|
|
chr6_-_39305203
|
0.467
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr3_-_18455207
|
0.465
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr10_+_112826774
|
0.462
|
NM_000681
|
ADRA2A
|
adrenergic, alpha-2A-, receptor
|
|
chr8_+_16929116
|
0.462
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr6_-_119137923
|
0.461
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr2_+_206847658
|
0.461
|
NM_020923
|
ZDBF2
|
zinc finger, DBF-type containing 2
|
|
chr8_-_93177057
|
0.458
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_+_124114985
|
0.457
|
NM_001126181
NM_006176
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr9_-_14900992
|
0.456
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr2_-_208342296
|
0.455
|
NM_003468
|
FZD5
|
frizzled homolog 5 (Drosophila)
|
|
chr14_+_66777578
|
0.436
|
NM_022474
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5)
|
|
chr10_+_103976132
|
0.431
|
NM_152310
|
ELOVL3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3
|
|
chr8_+_77756061
|
0.428
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr13_+_72199981
|
0.424
|
NM_024808
|
C13orf34
|
chromosome 13 open reading frame 34
|
|
chr11_-_83661878
|
0.423
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr2_+_135392857
|
0.416
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr14_+_72594940
|
0.414
|
NM_021239
|
RBM25
|
RNA binding motif protein 25
|
|
chr20_-_17897067
|
0.413
|
NM_014426
|
SNX5
|
sorting nexin 5
|
|
chr18_+_65219114
|
0.406
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chr6_+_106653429
|
0.399
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr3_+_113287864
|
0.390
|
NM_001171747
NM_024616
|
C3orf52
|
chromosome 3 open reading frame 52
|
|
chr17_-_18526296
|
0.387
|
NM_001145045
|
ZNF286B
|
zinc finger protein 286B
|
|
chr17_-_10362583
|
0.382
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr17_+_1905104
|
0.382
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr3_-_140148671
|
0.377
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr8_+_79590890
|
0.368
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr4_+_144477432
|
0.366
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr16_+_24648446
|
0.362
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr4_-_21559471
|
0.355
|
NM_147182
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr6_-_24827265
|
0.354
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr6_+_105511615
|
0.346
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr17_-_44010543
|
0.339
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr5_+_82803248
|
0.338
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr1_-_214963279
|
0.337
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr12_-_46499834
|
0.333
|
NM_001098416
NM_015401
|
HDAC7
|
histone deacetylase 7
|
|
chr8_-_93181091
|
0.332
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_68037378
|
0.324
|
NM_001193502
|
TCF24
|
transcription factor 24
|
|
chr11_-_16380967
|
0.323
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr16_+_87021379
|
0.323
|
NM_001127464
|
ZNF469
|
zinc finger protein 469
|
|
chr16_-_15889947
|
0.315
|
NM_144600
|
C16orf63
|
chromosome 16 open reading frame 63
|
|
chr14_+_101855836
|
0.314
|
NM_018335
|
ZNF839
|
zinc finger protein 839
|
|
chr5_+_162797149
|
0.314
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr4_-_13155135
|
0.313
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr1_-_92124201
|
0.313
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr6_-_134680888
|
0.312
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr20_+_43028533
|
0.310
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr20_-_17897384
|
0.308
|
NM_152227
|
SNX5
|
sorting nexin 5
|
|
chr22_+_43939193
|
0.307
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr6_+_106640881
|
0.307
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr14_+_38805251
|
0.302
|
NM_203356
|
CTAGE5
|
CTAGE family, member 5
|
|
chr6_+_15354278
|
0.300
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr2_+_101680594
|
0.288
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr14_+_76634323
|
0.287
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr1_-_85816520
|
0.284
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr16_-_23068010
|
0.283
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr11_-_110088330
|
0.282
|
NM_020809
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr13_-_79813042
|
0.275
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr6_-_119079712
|
0.275
|
NM_001042475
NM_206921
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr2_+_66516020
|
0.274
|
NM_002398
|
MEIS1
|
Meis homeobox 1
|
|
chr2_+_202607554
|
0.272
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr1_-_148936123
|
0.272
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr3_-_87122936
|
0.271
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr17_+_24742068
|
0.271
|
NM_020791
|
TAOK1
|
TAO kinase 1
|
|
chr3_-_66633534
|
0.269
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr16_-_74024750
|
0.267
|
NM_006324
|
CFDP1
|
craniofacial development protein 1
|
|
chr2_+_172658453
|
0.264
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr4_-_21308415
|
0.260
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr2_-_100305626
|
0.257
|
NM_198461
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2
|
|
chr14_-_34252695
|
0.257
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr17_+_76133206
|
0.256
|
NM_001163034
NM_020761
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr12_+_12829807
|
0.255
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr5_-_136862786
|
0.250
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr11_-_85015961
|
0.249
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr4_-_185632610
|
0.249
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr3_+_213535
|
0.248
|
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr20_+_30871357
|
0.247
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr3_+_148609841
|
0.242
|
NM_003412
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr4_-_78959532
|
0.242
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr14_-_38709161
|
0.241
|
NM_001079537
NM_177452
|
TRAPPC6B
|
trafficking protein particle complex 6B
|
|
chr7_-_97719372
|
0.240
|
NM_015395
|
TECPR1
|
tectonin beta-propeller repeat containing 1
|
|
chrX_-_110400406
|
0.239
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr15_-_68842799
|
0.237
|
NM_018003
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr10_-_101979332
|
0.235
|
NM_001278
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase
|
|
chr1_+_28458546
|
0.235
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr11_+_32870935
|
0.235
|
NM_001076786
|
QSER1
|
glutamine and serine rich 1
|
|
chr6_-_116681863
|
0.234
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr2_-_219633481
|
0.232
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr2_-_227737518
|
0.228
|
NM_000092
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr17_-_44026042
|
0.226
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chrX_-_99551926
|
0.224
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr1_-_92144146
|
0.222
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr4_+_154606900
|
0.221
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr2_+_5750229
|
0.219
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr6_+_121798443
|
0.213
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr1_+_178190493
|
0.211
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr18_-_802268
|
0.208
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr18_+_53865455
|
0.208
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr20_+_13150417
|
0.205
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr3_+_50687675
|
0.204
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr2_+_115635982
|
0.202
|
NM_001178034
NM_001004360
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr1_-_215377718
|
0.197
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr12_-_87498225
|
0.195
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr11_+_86426518
|
0.193
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr2_+_60836858
|
0.192
|
NM_022894
|
PAPOLG
|
poly(A) polymerase gamma
|
|
chrX_+_122821217
|
0.189
|
NM_001167
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr15_-_46725087
|
0.188
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr5_-_41546384
|
0.187
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr12_+_13088551
|
0.185
|
NM_020853
|
KIAA1467
|
KIAA1467
|
|
chr10_+_112247614
|
0.184
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr12_+_12769985
|
0.182
|
NM_001130415
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr6_-_64087806
|
0.181
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr5_+_56507029
|
0.180
|
NM_001127235
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr5_+_140194152
|
0.180
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr1_+_78729315
|
0.178
|
NM_000959
NM_001039585
|
PTGFR
|
prostaglandin F receptor (FP)
|
|
chr16_-_87535106
|
0.177
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr3_-_88281705
|
0.177
|
NM_001195308
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr10_+_11246934
|
0.177
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_+_121642204
|
0.176
|
NM_007190
|
SEC23IP
|
SEC23 interacting protein
|
|
chr4_+_166468266
|
0.175
|
NM_001017369
NM_006745
|
SC4MOL
|
sterol-C4-methyl oxidase-like
|
|
chr14_-_36059084
|
0.174
|
NM_001079668
|
NKX2-1
|
NK2 homeobox 1
|
|
chr14_-_38971370
|
0.174
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr14_-_36058648
|
0.173
|
NM_003317
|
NKX2-1
|
NK2 homeobox 1
|
|
chr18_-_30057512
|
0.171
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr11_-_16387001
|
0.170
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr18_-_30056431
|
0.168
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr3_+_116824840
|
0.168
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr11_-_46569489
|
0.167
|
NM_017749
|
AMBRA1
|
autophagy/beclin-1 regulator 1
|
|
chr12_+_120635021
|
0.167
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr3_-_71857097
|
0.164
|
NM_001134651
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr18_+_3441589
|
0.164
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr17_+_15543615
|
0.164
|
NM_001130842
NM_020652
|
ZNF286A
|
zinc finger protein 286A
|
|
chr1_-_215329569
|
0.163
|
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr20_+_36867708
|
0.162
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr16_-_51138306
|
0.161
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr17_-_38014930
|
0.160
|
NM_178126
|
FAM134C
|
family with sequence similarity 134, member C
|
|
chr5_-_107034484
|
0.160
|
NM_001962
|
EFNA5
|
ephrin-A5
|
|
chr2_-_55698217
|
0.160
|
NM_001122964
NM_020463
|
SMEK2
|
SMEK homolog 2, suppressor of mek1 (Dictyostelium)
|
|
chr2_-_171725604
|
0.159
|
NM_012290
|
TLK1
|
tousled-like kinase 1
|
|
chr11_+_22171150
|
0.158
|
NM_001142649
NM_213599
|
ANO5
|
anoctamin 5
|
|
chr18_+_3437607
|
0.158
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr2_-_20886293
|
0.158
|
NM_021925
|
C2orf43
|
chromosome 2 open reading frame 43
|
|
chr4_+_41631862
|
0.158
|
NM_018126
|
TMEM33
|
transmembrane protein 33
|
|
chr1_+_234625338
|
0.157
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr12_-_69289889
|
0.154
|
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr11_+_93916653
|
0.154
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr20_+_48781457
|
0.152
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr19_+_42261147
|
0.150
|
NM_144689
|
ZNF420
|
zinc finger protein 420
|