|
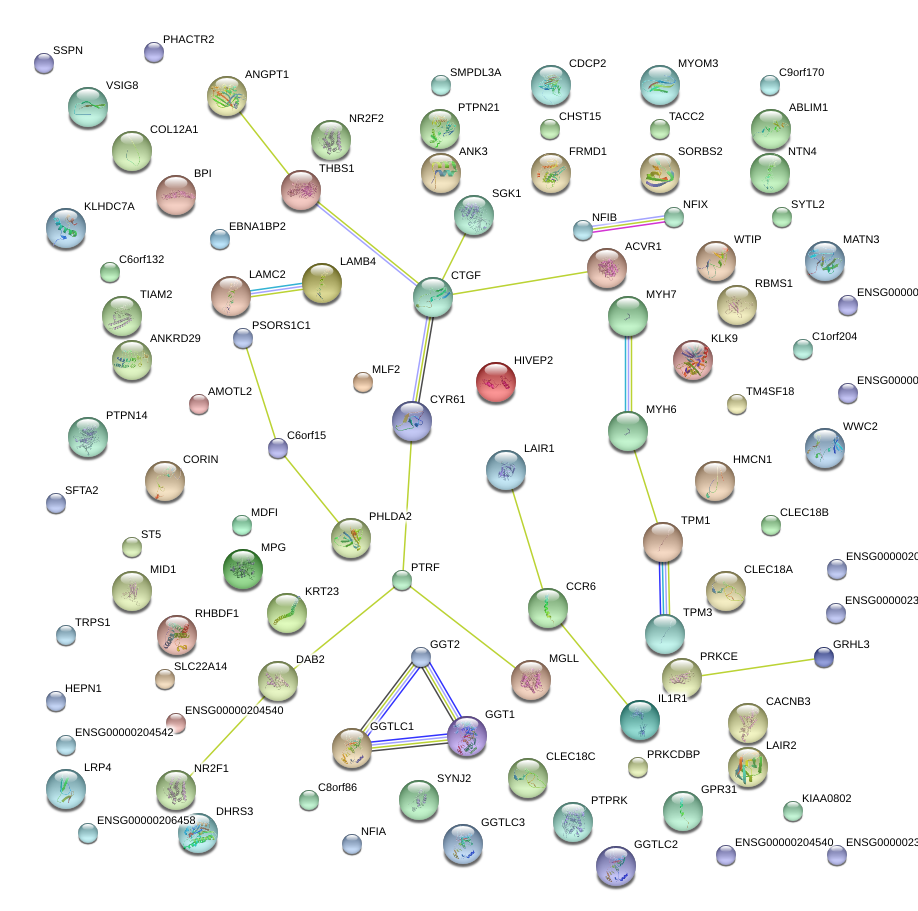
chr6_-_132314004
|
6.100
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr4_+_184257337
|
5.144
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr1_-_212791597
|
5.071
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr1_-_212791467
|
5.013
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr10_-_116382089
|
4.939
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr3_-_135575925
|
4.878
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr11_-_46896484
|
4.121
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr1_-_43509187
|
4.108
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chrX_-_10548458
|
3.887
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr10_+_123862430
|
3.780
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr2_-_160972606
|
3.694
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr6_-_128883125
|
3.586
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr15_+_61121806
|
3.320
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr12_-_94708641
|
3.201
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr15_+_37660568
|
3.060
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr6_+_41712907
|
3.039
|
|
MDFI
|
MyoD family inhibitor
|
|
chr10_-_61819493
|
3.014
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr5_-_39460687
|
2.964
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr14_-_88090641
|
2.959
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr1_+_24522116
|
2.909
|
NM_021180
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr1_-_12599934
|
2.824
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr1_+_18680010
|
2.741
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr11_-_6298284
|
2.725
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr1_-_24311212
|
2.696
|
NM_152372
|
MYOM3
|
myomesin family, member 3
|
|
chr16_-_62573
|
2.664
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr19_+_39664706
|
2.648
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr6_+_167445284
|
2.600
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr11_-_8789457
|
2.591
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr15_+_61122139
|
2.562
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr18_-_19496590
|
2.500
|
NM_173505
|
ANKRD29
|
ankyrin repeat domain 29
|
|
chr12_+_47498778
|
2.468
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr19_-_56204615
|
2.452
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr9_+_88953378
|
2.416
|
NM_001001709
|
C9orf170
|
chromosome 9 open reading frame 170
|
|
chr12_+_26239755
|
2.416
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr17_-_36347197
|
2.351
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr6_-_75972286
|
2.326
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr6_+_123152044
|
2.227
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr11_-_2907169
|
2.225
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr2_-_158439868
|
2.222
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr5_-_39460787
|
2.217
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr17_-_37828643
|
2.212
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr20_-_23915431
|
2.170
|
NM_178312
|
GGTLC1
|
gamma-glutamyltransferase light chain 1
|
|
chr10_-_125843092
|
2.161
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr6_-_168222645
|
2.148
|
NM_024919
|
FRMD1
|
FERM domain containing 1
|
|
chr11_-_8788765
|
2.148
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr19_-_59568222
|
2.132
|
|
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1
|
|
chr3_-_129023850
|
2.130
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr4_-_186970366
|
2.089
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr19_-_59568340
|
2.071
|
|
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1
|
|
chr1_-_158098962
|
1.923
|
NM_001013661
|
VSIG8
|
V-set and immunoglobulin domain containing 8
|
|
chr1_+_85819004
|
1.921
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr16_+_68765428
|
1.919
|
NM_173619
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr6_+_155453114
|
1.910
|
NM_012454
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr2_+_45732416
|
1.896
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr11_+_124294355
|
1.893
|
NM_001037558
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1
|
|
chr20_+_36365965
|
1.865
|
NM_001725
|
BPI
|
bactericidal/permeability-increasing protein
|
|
chr1_-_54392030
|
1.856
|
NM_201546
|
CDCP2
|
CUB domain containing protein 2
|
|
chr6_-_167491305
|
1.843
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr6_-_134538762
|
1.832
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_181422016
|
1.817
|
|
LAMC2
|
laminin, gamma 2
|
|
chr8_-_38505331
|
1.809
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr6_-_143307976
|
1.778
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr2_-_20075929
|
1.753
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr6_-_31007905
|
1.742
|
NM_205854
|
SFTA2
|
surfactant associated 2
|
|
chr6_-_31188310
|
1.739
|
NM_014070
|
C6orf15
|
chromosome 6 open reading frame 15
|
|
chr2_-_20075889
|
1.727
|
|
MATN3
|
matrilin 3
|
|
chr7_-_107558021
|
1.720
|
NM_007356
|
LAMB4
|
laminin, beta 4
|
|
chr2_+_102125616
|
1.719
|
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr1_+_181421984
|
1.689
|
|
LAMC2
|
laminin, gamma 2
|
|
chr22_+_21318779
|
1.662
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr6_-_42218692
|
1.656
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr3_+_38322430
|
1.651
|
NM_004803
|
SLC22A14
|
solute carrier family 22, member 14
|
|
chr3_-_150533948
|
1.641
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr11_-_85107569
|
1.641
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr19_-_59568483
|
1.638
|
NM_002287
NM_021706
|
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1
|
|
chr11_-_85108030
|
1.637
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr15_+_94670160
|
1.627
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr14_-_22947320
|
1.622
|
NM_002471
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha
|
|
chr18_+_8707368
|
1.607
|
NM_015210
|
KIAA0802
|
KIAA0802
|
|
chr8_-_108579270
|
1.587
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr1_+_183970093
|
1.582
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr6_+_144040751
|
1.556
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr8_-_116750427
|
1.488
|
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr6_+_158322875
|
1.486
|
NM_003898
|
SYNJ2
|
synaptojanin 2
|
|
chr1_+_61320213
|
1.465
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr6_+_31190505
|
1.459
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr3_+_138020550
|
1.457
|
NM_001097599
NM_001097600
NM_025246
|
TMEM22
|
transmembrane protein 22
|
|
chr14_-_22516271
|
1.425
|
NM_198086
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr5_-_37871685
|
1.421
|
NM_001190468
NM_001190469
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr1_+_181421796
|
1.421
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr1_-_16355013
|
1.416
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr8_-_38444398
|
1.403
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr1_-_201583989
|
1.402
|
|
FMOD
|
fibromodulin
|
|
chr4_-_186815116
|
1.400
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr7_+_134114701
|
1.398
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr7_+_134114912
|
1.380
|
|
CALD1
|
caldesmon 1
|
|
chr16_-_73012740
|
1.366
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr6_+_158322916
|
1.364
|
|
SYNJ2
|
synaptojanin 2
|
|
chr1_+_228949752
|
1.355
|
NM_006615
NM_016452
|
CAPN9
|
calpain 9
|
|
chr6_+_116956868
|
1.347
|
NM_153036
|
FAM26D
|
family with sequence similarity 26, member D
|
|
chr2_+_113546365
|
1.338
|
NM_032556
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chr1_-_181889070
|
1.326
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr3_+_9919295
|
1.313
|
NM_153483
|
IL17RE
|
interleukin 17 receptor E
|
|
chr2_+_149118791
|
1.296
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr5_-_39461054
|
1.290
|
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr6_-_75972473
|
1.278
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr5_-_148422799
|
1.269
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr12_-_47464143
|
1.229
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chr1_+_26358097
|
1.212
|
NM_024869
|
GRRP1
|
glycine/arginine rich protein 1
|
|
chr15_-_39973566
|
1.201
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr6_+_158322904
|
1.200
|
|
SYNJ2
|
synaptojanin 2
|
|
chr1_-_17179694
|
1.195
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr12_-_53099277
|
1.163
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr5_+_34723420
|
1.158
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr14_-_22515725
|
1.143
|
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr18_+_19947314
|
1.141
|
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr5_-_138767654
|
1.121
|
NM_194296
|
SPATA24
|
spermatogenesis associated 24
|
|
chr7_+_134114957
|
1.114
|
|
CALD1
|
caldesmon 1
|
|
chr9_+_115970599
|
1.113
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr2_+_151922350
|
1.105
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr5_+_131657964
|
1.105
|
NM_003059
|
SLC22A4
|
solute carrier family 22 (organic cation/ergothioneine transporter), member 4
|
|
chr2_+_37282516
|
1.104
|
|
LOC100505876
|
hypothetical LOC100505876
|
|
chr7_+_115952279
|
1.103
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr4_-_152368492
|
1.101
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr4_+_152417774
|
1.098
|
NM_183375
|
PRSS48
|
protease, serine, 48
|
|
chr17_+_71229110
|
1.092
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr16_-_21344158
|
1.090
|
NM_130464
|
NPIPL3
|
nuclear pore complex interacting protein-like 3
|
|
chr22_-_20419975
|
1.088
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr3_+_191588354
|
1.086
|
NM_006580
|
CLDN16
|
claudin 16
|
|
chr2_-_192419896
|
1.067
|
|
SDPR
|
serum deprivation response
|
|
chr5_+_76150637
|
1.066
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr11_-_124311106
|
1.065
|
|
HEPACAM
|
hepatic and glial cell adhesion molecule
|
|
chr12_-_53099269
|
1.053
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr1_-_199704773
|
1.048
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr17_+_71229191
|
1.020
|
|
ITGB4
|
integrin, beta 4
|
|
chr16_+_24174975
|
1.016
|
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr6_-_31789824
|
1.003
|
|
|
|
|
chr8_-_125785611
|
0.999
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr11_-_9182356
|
0.995
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr1_+_20269239
|
0.983
|
NM_000929
|
PLA2G5
|
phospholipase A2, group V
|
|
chr17_+_71229151
|
0.981
|
|
ITGB4
|
integrin, beta 4
|
|
chr3_+_9919511
|
0.977
|
NM_001193380
NM_153480
|
IL17RE
|
interleukin 17 receptor E
|
|
chr6_-_134538526
|
0.970
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_-_53099203
|
0.954
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr10_-_17699309
|
0.935
|
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A
|
|
chr9_+_117955890
|
0.935
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr5_+_140848905
|
0.931
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr5_+_38882017
|
0.926
|
|
OSMR
|
oncostatin M receptor
|
|
chr3_-_115825742
|
0.925
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr16_+_15435825
|
0.903
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr1_-_149698516
|
0.902
|
NM_001194937
NM_001194938
NM_015100
NM_145796
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr13_+_109936080
|
0.900
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr9_+_94776538
|
0.887
|
NM_033086
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr2_+_95303947
|
0.885
|
|
PROM2
|
prominin 2
|
|
chr19_+_55383249
|
0.884
|
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr5_-_38631263
|
0.875
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr19_-_56147971
|
0.871
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr2_-_75641587
|
0.867
|
NM_001135032
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr2_-_75641535
|
0.865
|
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr5_-_159672060
|
0.863
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr3_+_125586241
|
0.850
|
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr3_-_113842553
|
0.847
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr5_-_159730196
|
0.844
|
NM_031908
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2
|
|
chrX_-_24241352
|
0.841
|
NM_001136233
|
FAM48B2
|
family with sequence similarity 48, member B2
|
|
chr7_+_115952325
|
0.837
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr3_-_113842237
|
0.836
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr4_-_184480572
|
0.829
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chr12_-_119172339
|
0.826
|
NM_025157
|
PXN
|
paxillin
|
|
chr15_-_97366313
|
0.817
|
NM_001102612
|
PGPEP1L
|
pyroglutamyl-peptidase I-like
|
|
chr12_-_78608873
|
0.813
|
NM_002583
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chr11_+_1812115
|
0.811
|
NM_138567
|
SYT8
|
synaptotagmin VIII
|
|
chr3_-_113841967
|
0.809
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_10307408
|
0.808
|
NM_001134941
NM_016362
|
GHRL
|
ghrelin/obestatin prepropeptide
|
|
chr1_+_242281199
|
0.804
|
|
ZNF238
|
zinc finger protein 238
|
|
chr11_-_65082705
|
0.803
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr5_-_37871349
|
0.800
|
NM_199231
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr19_+_522260
|
0.799
|
NM_198591
|
BSG
|
basigin (Ok blood group)
|
|
chr10_+_99334069
|
0.795
|
NM_001134670
NM_138413
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr16_+_22432344
|
0.792
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chr17_+_46267603
|
0.792
|
NM_175575
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2
|
|
chr6_+_31910623
|
0.791
|
NM_001040437
NM_001040438
|
C6orf48
|
chromosome 6 open reading frame 48
|
|
chr11_+_57123927
|
0.791
|
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr3_-_113842350
|
0.788
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr6_-_42017337
|
0.783
|
NM_001136125
NM_001760
|
CCND3
|
cyclin D3
|
|
chr14_-_22521544
|
0.783
|
NM_032876
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr19_-_51666503
|
0.782
|
|
PNMAL1
|
PNMA-like 1
|
|
chr2_-_233349469
|
0.779
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr10_+_102782151
|
0.777
|
|
SFXN3
|
sideroflexin 3
|
|
chr3_-_113842133
|
0.776
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr2_+_95303922
|
0.774
|
NM_001165977
NM_001165978
NM_144707
|
PROM2
|
prominin 2
|
|
chr11_-_124311512
|
0.774
|
NM_152722
|
HEPACAM
|
hepatic and glial cell adhesion molecule
|
|
chr21_+_43024936
|
0.771
|
|
PDE9A
|
phosphodiesterase 9A
|
|
chr20_-_55719946
|
0.766
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr1_+_216585298
|
0.764
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr11_+_46323575
|
0.756
|
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr15_+_73728402
|
0.754
|
NM_153271
|
SNX33
|
sorting nexin 33
|
|
chr6_+_108987703
|
0.754
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr3_-_113842255
|
0.753
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chrX_+_30169977
|
0.751
|
NM_002367
|
MAGEB4
|
melanoma antigen family B, 4
|
|
chr17_-_3445873
|
0.751
|
NM_080705
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1
|