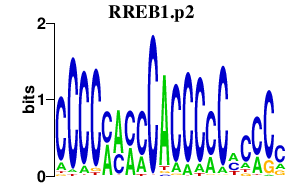
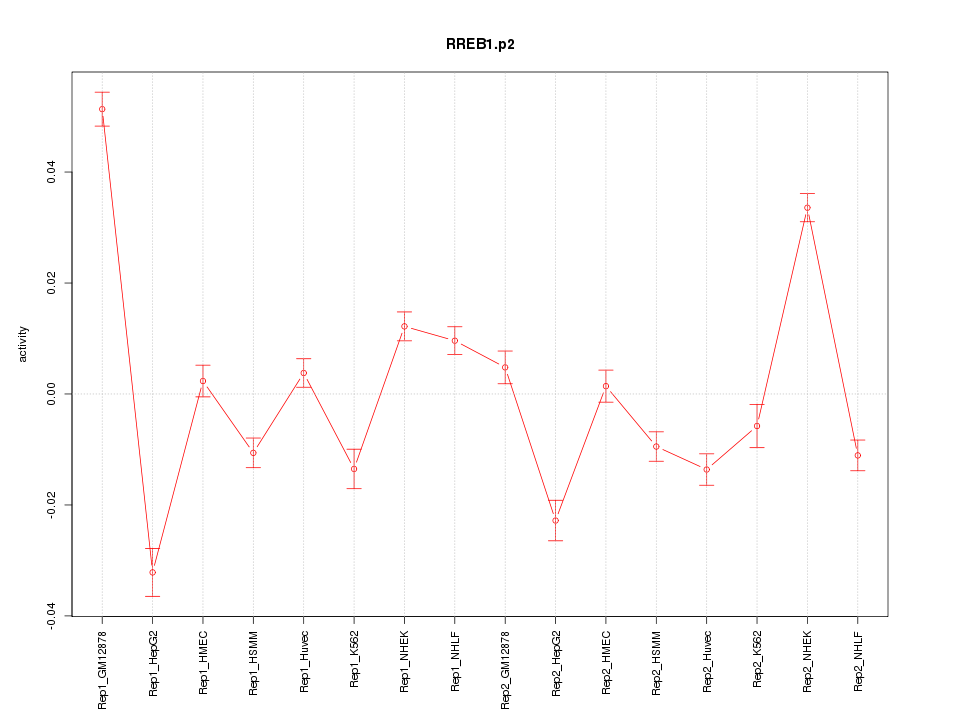
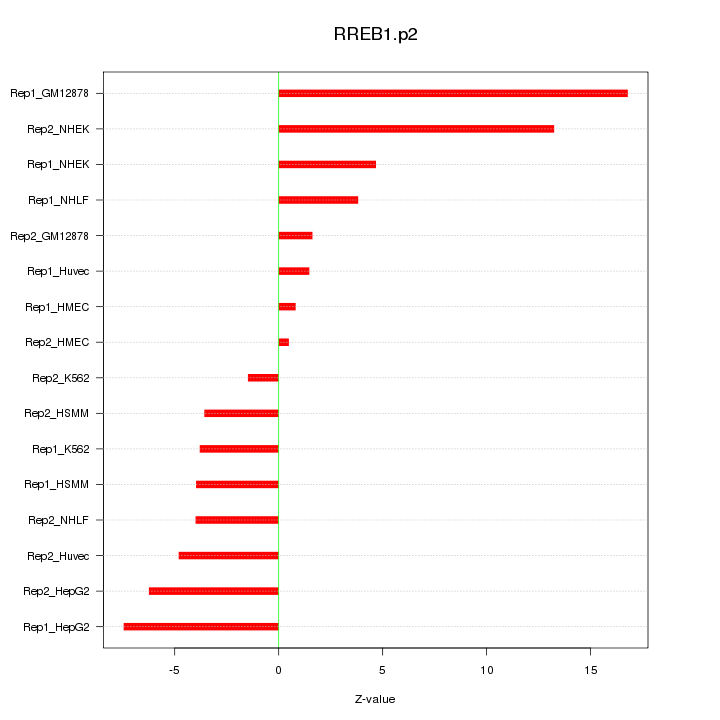
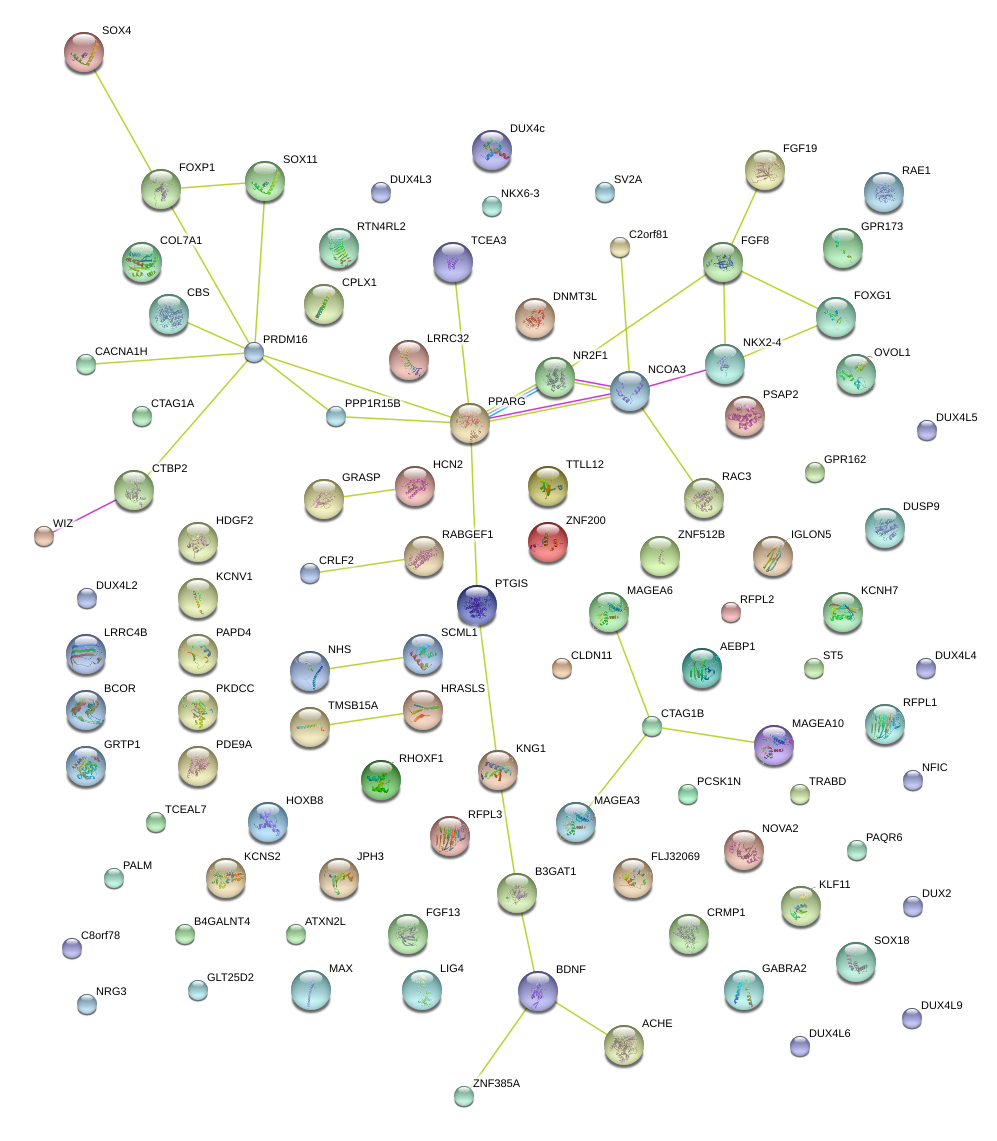
|
chrX_+_17665483
|
13.433
|
NM_001037535
NM_001037536
NM_001037540
NM_006746
|
SCML1
|
sex comb on midleg-like 1 (Drosophila)
|
|
chr16_-_3225360
|
12.578
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr19_-_55914837
|
11.479
|
|
|
|
|
chr4_-_809879
|
10.811
|
NM_006651
|
CPLX1
|
complexin 1
|
|
chr20_+_55359551
|
10.456
|
NM_003610
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr3_+_187917791
|
10.069
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr20_+_55359706
|
9.929
|
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr21_+_42946719
|
9.555
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr16_-_3225070
|
9.264
|
NM_001145447
NM_003454
|
ZNF200
|
zinc finger protein 200
|
|
chr19_-_51168396
|
8.642
|
NM_002516
|
NOVA2
|
neuro-oncological ventral antigen 2
|
|
chr7_+_44110527
|
7.867
|
|
AEBP1
|
AE binding protein 1
|
|
chr11_-_133786935
|
7.729
|
NM_018644
NM_054025
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P)
|
|
chr7_+_44110470
|
7.708
|
NM_001129
|
AEBP1
|
AE binding protein 1
|
|
chr4_-_46086528
|
7.474
|
NM_000807
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2
|
|
chr1_-_202647526
|
7.158
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory (inhibitor) subunit 15B
|
|
chr16_+_86193993
|
7.002
|
NM_020655
|
JPH3
|
junctophilin 3
|
|
chr19_+_3317589
|
6.886
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr22_+_48966394
|
6.858
|
|
TRABD
|
TraB domain containing
|
|
chr22_+_48966480
|
6.731
|
NM_025204
|
TRABD
|
TraB domain containing
|
|
chr4_+_191235959
|
6.705
|
NM_001177376
|
DUX4L4
|
double homeobox 4 like 4
|
|
chr5_+_78943998
|
6.685
|
NM_001114393
NM_001114394
NM_173797
|
PAPD4
|
PAP associated domain containing 4
|
|
chr17_+_77582774
|
6.469
|
NM_005052
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3)
|
|
chr5_+_78944052
|
6.162
|
|
PAPD4
|
PAP associated domain containing 4
|
|
chr3_+_12304332
|
6.097
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr12_+_6807831
|
5.948
|
NM_014262
|
LEPREL2
|
leprecan-like 2
|
|
chr19_+_540849
|
5.724
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr2_+_42128521
|
5.655
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr5_+_78944151
|
5.529
|
|
PAPD4
|
PAP associated domain containing 4
|
|
chrX_-_119133802
|
5.477
|
NM_139282
|
RHOXF1
|
Rhox homeobox family, member 1
|
|
chr14_-_64638809
|
5.383
|
|
MAX
|
MYC associated factor X
|
|
chr19_+_3317575
|
5.267
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr1_-_154318412
|
5.177
|
NM_001093725
|
MEX3A
|
mex-3 homolog A (C. elegans)
|
|
chr20_-_47618059
|
5.141
|
NM_000961
|
PTGIS
|
prostaglandin I2 (prostacyclin) synthase
|
|
chr2_-_163403274
|
5.051
|
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7
|
|
chr13_-_107665871
|
5.046
|
NM_206937
|
LIG4
|
ligase IV, DNA, ATP-dependent
|
|
chr22_-_41913002
|
4.947
|
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12
|
|
chr4_+_191245840
|
4.908
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr16_+_28741873
|
4.818
|
|
ATXN2L
|
ataxin 2-like
|
|
chr4_+_191232653
|
4.695
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chrX_+_53095230
|
4.650
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chrX_-_101658297
|
4.533
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr16_+_28741908
|
4.496
|
NM_007245
NM_145714
NM_148414
NM_148415
NM_148416
|
ATXN2L
|
ataxin 2-like
|
|
chr12_-_53071285
|
4.466
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr19_+_3317536
|
4.447
|
NM_005597
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr16_+_28741861
|
4.437
|
|
ATXN2L
|
ataxin 2-like
|
|
chr22_-_41913076
|
4.437
|
NM_015140
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12
|
|
chr22_-_30929463
|
4.385
|
NM_001098527
|
RFPL2
|
ret finger protein-like 2
|
|
chr19_+_4442610
|
4.377
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr2_+_5750229
|
4.368
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr21_-_43368963
|
4.215
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chrX_-_1291526
|
4.134
|
NM_001012288
NM_022148
|
CRLF2
|
cytokine receptor-like factor 2
|
|
chr22_-_41913075
|
4.131
|
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12
|
|
chr7_+_65731364
|
4.052
|
|
KCTD7
|
potassium channel tetramerisation domain containing 7
|
|
chr11_+_65311104
|
3.977
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr21_-_43369492
|
3.963
|
NM_001178008
NM_001178009
|
CBS
|
cystathionine-beta-synthase
|
|
chr3_+_171621721
|
3.951
|
NM_001185056
|
CLDN11
|
claudin 11
|
|
chrX_-_151057615
|
3.912
|
NM_001011543
NM_021048
|
MAGEA10
|
melanoma antigen family A, 10
|
|
chr4_+_191226073
|
3.870
|
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
|
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
|
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
|
|
chr19_+_659766
|
3.835
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chrX_+_152561086
|
3.808
|
NM_001395
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr20_-_21326046
|
3.802
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr1_-_23623713
|
3.663
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr16_+_28742066
|
3.652
|
|
ATXN2L
|
ataxin 2-like
|
|
chr4_-_5941043
|
3.645
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr10_-_103525646
|
3.634
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr11_-_76059435
|
3.630
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr10_+_135333667
|
3.588
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr11_-_27678551
|
3.560
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chrX_-_137621492
|
3.519
|
NM_004114
|
FGF13
|
fibroblast growth factor 13
|
|
chr3_+_194441586
|
3.509
|
NM_020386
|
HRASLS
|
HRAS-like suppressor
|
|
chr5_+_92946348
|
3.492
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr1_-_148155986
|
3.485
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr4_-_5941167
|
3.479
|
NM_001313
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr1_-_154484341
|
3.448
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr19_-_15421761
|
3.377
|
NM_021241
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr1_+_1066138
|
3.364
|
|
|
|
|
chr11_+_65311142
|
3.361
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr22_+_49459935
|
3.347
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr3_-_71715431
|
3.329
|
NM_001012505
NM_032682
|
FOXP1
|
forkhead box P1
|
|
chr11_+_56984702
|
3.291
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chrX_+_153466611
|
3.291
|
NM_001327
NM_139250
|
CTAG1B
CTAG1A
|
cancer/testis antigen 1B
cancer/testis antigen 1A
|
|
chrX_-_48578878
|
3.259
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr4_+_191242540
|
3.257
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr7_-_100331417
|
3.251
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr17_-_44047299
|
3.242
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr11_+_359723
|
3.213
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr20_-_26137755
|
3.177
|
|
LOC284801
|
hypothetical protein LOC284801
|
|
chr4_+_191239247
|
3.173
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr16_+_1143241
|
3.170
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr1_+_2975590
|
3.168
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr10_+_135343649
|
3.164
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr20_-_62151216
|
3.150
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr1_-_182273114
|
3.143
|
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr13_-_113066202
|
3.127
|
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr14_+_28305922
|
3.103
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chrX_+_102471769
|
3.071
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chrX_-_39841599
|
3.063
|
NM_001123385
NM_017745
|
BCOR
|
BCL6 corepressor
|
|
chr21_-_44506526
|
3.052
|
NM_013369
NM_175867
|
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like
|
|
chr11_-_8695974
|
3.045
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr2_-_163403485
|
3.036
|
NM_033272
NM_173162
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7
|
|
chr11_-_76058542
|
3.031
|
NM_005512
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr20_-_62071584
|
3.023
|
NM_020713
|
ZNF512B
|
zinc finger protein 512B
|
|
chr3_-_48607596
|
3.017
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr8_-_41624030
|
3.015
|
NM_152568
|
NKX6-3
|
NK6 homeobox 3
|
|
chr16_+_1143738
|
3.005
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr10_-_126839000
|
2.994
|
|
CTBP2
|
C-terminal binding protein 2
|
|
chr11_-_69228286
|
2.948
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr10_+_83625049
|
2.941
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr19_-_55763091
|
2.928
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr19_+_56506907
|
2.925
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr11_-_27679022
|
2.923
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr8_-_125252884
|
2.910
|
|
C8orf78
|
chromosome 8 open reading frame 78
|
|
chr8_+_99508425
|
2.905
|
NM_020697
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2
|
|
chr17_-_60208083
|
2.891
|
|
LOC146880
|
hypothetical LOC146880
|
|
chr4_+_191229360
|
2.865
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr2_+_10101081
|
2.848
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr8_-_111056099
|
2.838
|
NM_014379
|
KCNV1
|
potassium channel, subfamily V, member 1
|
|
chr12_+_50686990
|
2.835
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chrX_+_151617900
|
2.810
|
NM_005363
NM_175868
|
MAGEA6
|
melanoma antigen family A, 6
|
|
chr20_+_48032918
|
2.797
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr15_-_54544531
|
2.791
|
NM_018365
|
MNS1
|
meiosis-specific nuclear structural 1
|
|
chr13_-_36392370
|
2.785
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr5_+_128823904
|
2.784
|
NM_133638
|
ADAMTS19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19
|
|
chr2_+_235525355
|
2.781
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr18_+_75261231
|
2.778
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr12_+_52665196
|
2.767
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr15_-_49174107
|
2.764
|
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3
|
|
chr9_-_115901157
|
2.763
|
NM_138424
|
KIF12
|
kinesin family member 12
|
|
chr11_-_6633427
|
2.755
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr17_+_26742903
|
2.738
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr4_+_3264509
|
2.717
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr19_+_40213431
|
2.682
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr19_+_38979502
|
2.669
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr17_-_74361975
|
2.662
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr6_-_48144344
|
2.657
|
NM_001013732
NM_207499
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr1_+_1940605
|
2.648
|
NM_000815
|
GABRD
|
gamma-aminobutyric acid (GABA) A receptor, delta
|
|
chr15_-_72952648
|
2.636
|
NM_005697
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr19_-_13478037
|
2.630
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr3_-_41978464
|
2.629
|
NM_017886
|
ULK4
|
unc-51-like kinase 4 (C. elegans)
|
|
chr16_+_86194075
|
2.602
|
|
JPH3
|
junctophilin 3
|
|
chr16_+_88517187
|
2.598
|
NM_006086
|
TUBB3
|
tubulin, beta 3
|
|
chrX_+_128501862
|
2.598
|
NM_000276
NM_001587
|
OCRL
|
oculocerebrorenal syndrome of Lowe
|
|
chr1_+_154318954
|
2.596
|
|
LMNA
|
lamin A/C
|
|
chr12_+_6808025
|
2.591
|
|
LEPREL2
|
leprecan-like 2
|
|
chr15_+_22644448
|
2.591
|
|
|
|
|
chrX_-_131918905
|
2.577
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr15_-_43267139
|
2.572
|
|
SHF
|
Src homology 2 domain containing F
|
|
chr12_-_21818881
|
2.567
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr3_+_213430
|
2.535
|
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chrX_-_148476903
|
2.525
|
NM_005365
NM_001080790
|
MAGEA9
MAGEA9B
|
melanoma antigen family A, 9
melanoma antigen family A, 9B
|
|
chr2_-_219881937
|
2.523
|
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chrX_-_152139267
|
2.518
|
NM_004988
|
MAGEA1
|
melanoma antigen family A, 1 (directs expression of antigen MZ2-E)
|
|
chr10_-_64248932
|
2.502
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr10_+_135330357
|
2.501
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chrX_-_151688850
|
2.486
|
NM_005362
|
MAGEA3
|
melanoma antigen family A, 3
|
|
chr2_+_242679507
|
2.480
|
|
LOC728323
|
hypothetical LOC728323
|
|
chr5_-_136862130
|
2.469
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chrX_-_135161185
|
2.464
|
NM_001173517
NM_024597
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr10_-_135000358
|
2.457
|
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr7_-_127833231
|
2.452
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr19_+_3831675
|
2.443
|
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chrX_+_148671394
|
2.434
|
NM_005365
NM_001080790
|
MAGEA9
MAGEA9B
|
melanoma antigen family A, 9
melanoma antigen family A, 9B
|
|
chr7_-_526005
|
2.429
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr11_-_88864061
|
2.422
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr13_-_107665130
|
2.413
|
NM_002312
|
LIG4
|
ligase IV, DNA, ATP-dependent
|
|
chr6_+_12120709
|
2.411
|
NM_002114
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1
|
|
chr3_-_159306214
|
2.410
|
|
|
|
|
chr3_+_112273352
|
2.404
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr3_+_50629593
|
2.392
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr14_-_20563698
|
2.391
|
|
NDRG2
|
NDRG family member 2
|
|
chr3_+_213535
|
2.379
|
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chrX_+_113724806
|
2.373
|
NM_000868
|
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chr11_-_118799058
|
2.368
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr14_+_102458745
|
2.358
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr14_+_68796621
|
2.358
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr19_+_57492236
|
2.338
|
NM_144684
|
ZNF480
|
zinc finger protein 480
|
|
chr1_-_75854281
|
2.333
|
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr22_+_47350781
|
2.328
|
NM_015381
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr14_-_20563722
|
2.320
|
NM_016250
NM_201535
NM_201536
NM_201537
|
NDRG2
|
NDRG family member 2
|
|
chr16_+_68516401
|
2.294
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr12_+_50271283
|
2.285
|
NM_014191
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr3_-_116348780
|
2.271
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_+_65731289
|
2.270
|
NM_001167961
NM_153033
|
KCTD7
|
potassium channel tetramerisation domain containing 7
|
|
chr17_+_58908152
|
2.259
|
NM_000789
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr11_-_2115077
|
2.252
|
|
|
|
|
chr19_+_522260
|
2.251
|
NM_198591
|
BSG
|
basigin (Ok blood group)
|
|
chr7_-_150129295
|
2.242
|
NM_014020
|
TMEM176B
|
transmembrane protein 176B
|
|
chr17_-_34014390
|
2.225
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr3_+_50629604
|
2.204
|
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chrX_-_71442488
|
2.203
|
NM_001144885
NM_001144887
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1
|
|
chr14_-_64638974
|
2.198
|
NM_002382
NM_145112
NM_145113
NM_145114
NM_145116
NM_197957
|
MAX
|
MYC associated factor X
|
|
chr3_+_129355006
|
2.193
|
|
EEFSEC
|
eukaryotic elongation factor, selenocysteine-tRNA-specific
|
|
chrX_-_47394937
|
2.188
|
NM_001114123
NM_005229
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr1_+_15123165
|
2.187
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr13_-_76499297
|
2.186
|
NM_012158
|
FBXL3
|
F-box and leucine-rich repeat protein 3
|
|
chr17_+_75366524
|
2.177
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr12_-_47679176
|
2.177
|
NM_015086
|
DDN
|
dendrin
|
|
chr19_-_5291700
|
2.170
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr19_-_15204767
|
2.169
|
NM_001142886
|
EPHX3
|
epoxide hydrolase 3
|
|
chr3_+_129354995
|
2.156
|
NM_021937
|
EEFSEC
|
eukaryotic elongation factor, selenocysteine-tRNA-specific
|