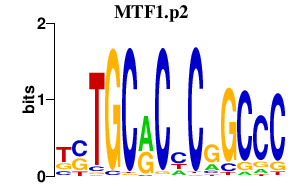
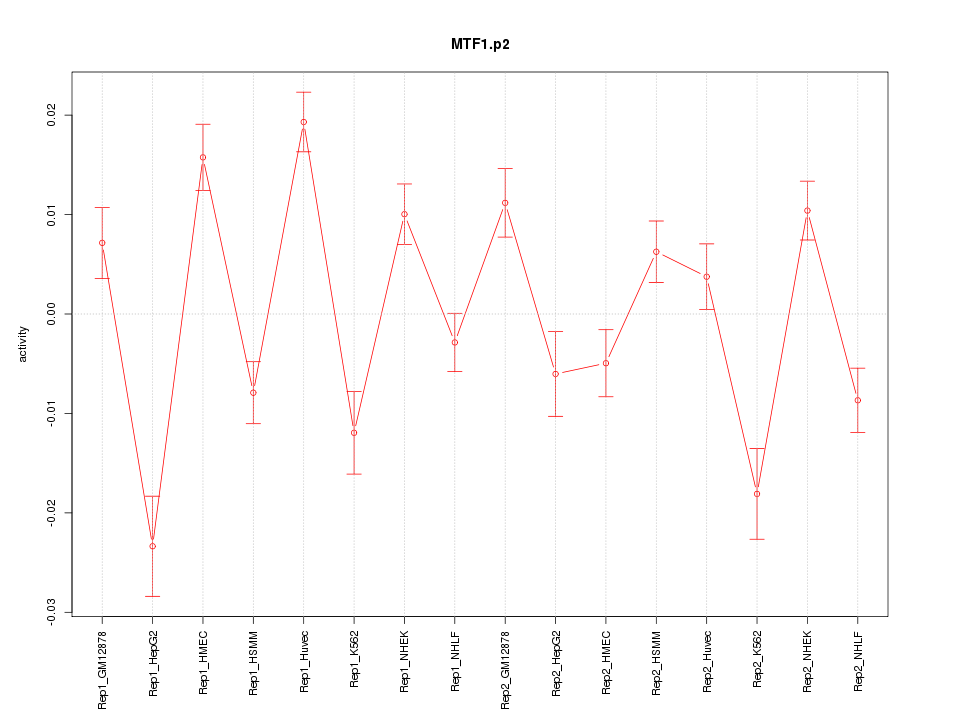
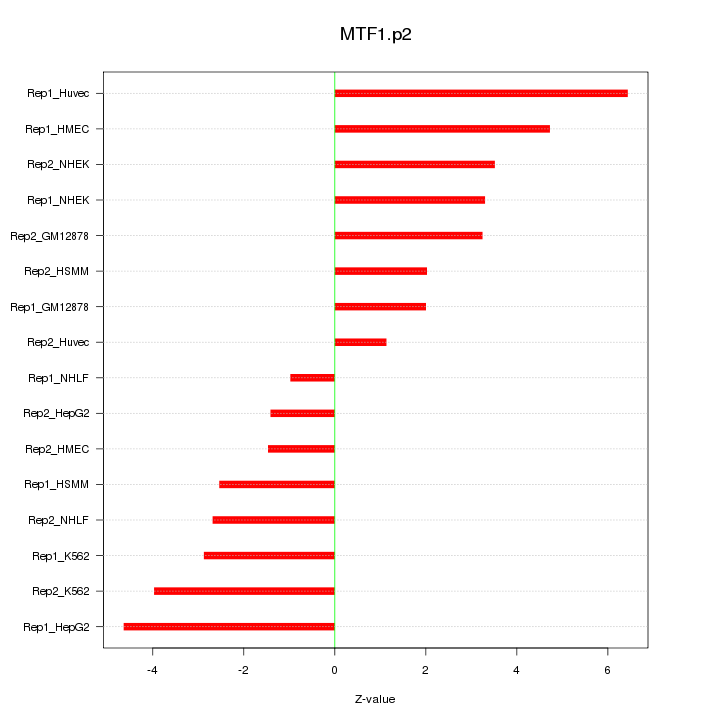
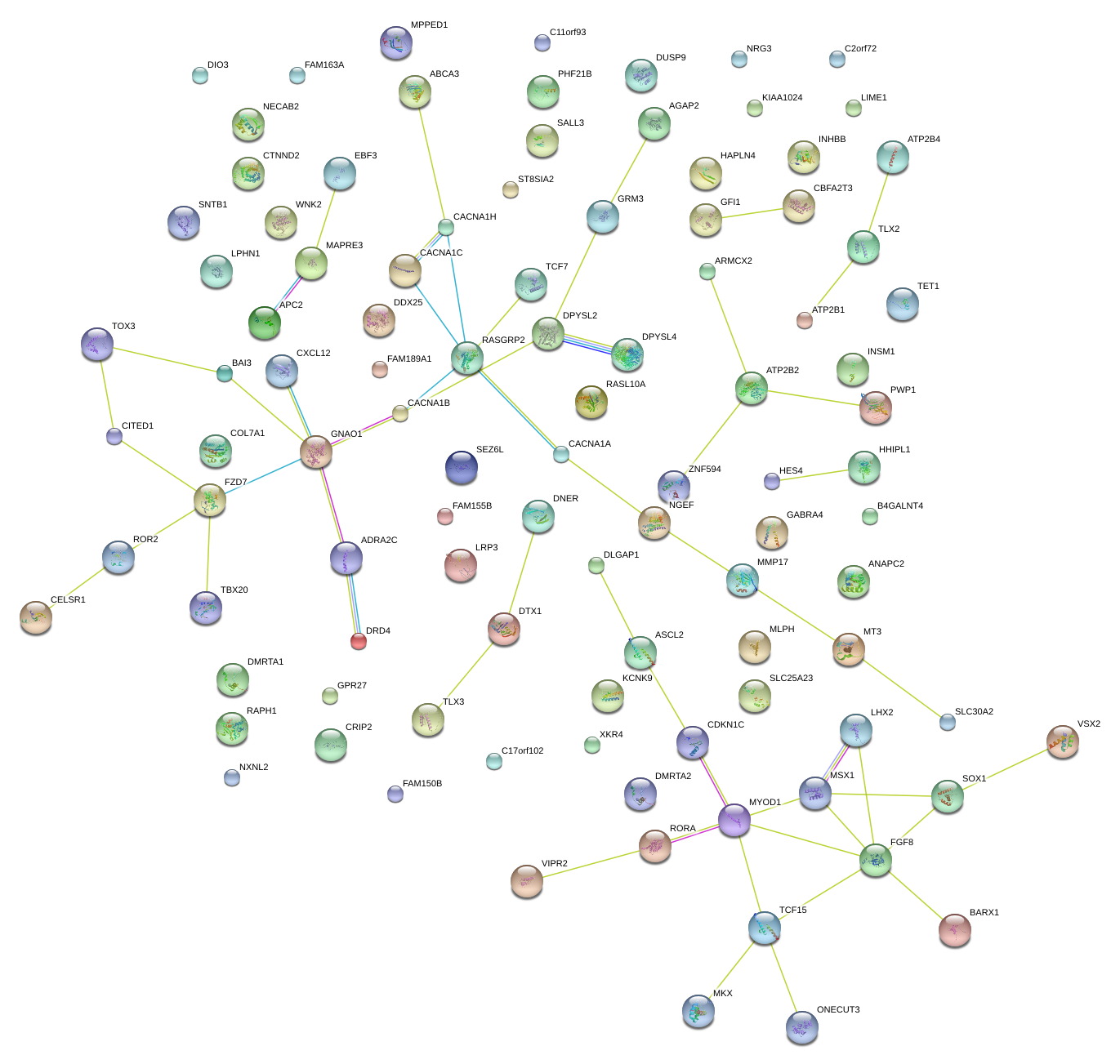
|
chr8_-_121893468
|
6.163
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr15_-_27650218
|
5.519
|
NM_015307
|
FAM189A1
|
family with sequence similarity 189, member A1
|
|
chr14_+_101097440
|
5.092
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chr8_-_121893449
|
4.817
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr5_-_11957127
|
4.781
|
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr9_+_90339835
|
4.669
|
NM_001161625
NM_145283
|
NXNL2
|
nucleoredoxin-like 2
|
|
chr8_+_56177567
|
4.544
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr18_+_74841262
|
4.410
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr10_-_103525646
|
4.167
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr5_-_11956964
|
4.036
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr19_+_1704661
|
4.031
|
NM_001080488
|
ONECUT3
|
one cut homeobox 3
|
|
chr8_-_121892877
|
3.791
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr8_-_121893196
|
3.789
|
|
|
|
|
chr8_-_140784416
|
3.752
|
NM_016601
|
KCNK9
|
potassium channel, subfamily K, member 9
|
|
chr11_-_2863536
|
3.607
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr9_+_139892049
|
3.567
|
NM_000718
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr10_+_83625049
|
3.529
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr16_-_87535106
|
3.501
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr2_+_74595073
|
3.461
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr16_-_51138306
|
3.398
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr9_-_93751900
|
3.259
|
NM_004560
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2
|
|
chr16_+_1143738
|
3.203
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chrX_+_68641802
|
3.176
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr16_+_55180767
|
3.061
|
NM_005954
|
MT3
|
metallothionein 3
|
|
chr3_-_62835601
|
3.047
|
|
|
|
|
chr2_+_238060602
|
2.998
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr11_+_125279481
|
2.957
|
NM_013264
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr13_+_111769913
|
2.940
|
NM_005986
|
SOX1
|
SRY (sex determining region Y)-box 1
|
|
chr8_+_121892719
|
2.808
|
|
|
|
|
chr19_-_14177980
|
2.806
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr9_-_95757365
|
2.721
|
NM_021570
|
BARX1
|
BARX homeobox 1
|
|
chr2_+_120820140
|
2.697
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr10_-_28074752
|
2.689
|
NM_173576
|
MKX
|
mohawk homeobox
|
|
chr10_-_131652364
|
2.686
|
|
EBF3
|
early B-cell factor 3
|
|
chr7_+_86111887
|
2.669
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr10_-_44200458
|
2.648
|
NM_000609
NM_001033886
NM_001178134
NM_199168
|
CXCL12
|
chemokine (C-X-C motif) ligand 12
|
|
chr1_-_92721633
|
2.648
|
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr2_-_233501069
|
2.627
|
NM_001114090
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr7_-_35260213
|
2.609
|
NM_001077653
NM_001166220
|
TBX20
|
T-box 20
|
|
chr9_+_125813849
|
2.607
|
|
LHX2
|
LIM homeobox 2
|
|
chr9_-_95757448
|
2.584
|
|
BARX1
|
BARX homeobox 1
|
|
chr10_-_135088065
|
2.546
|
NM_001012508
|
SPRN
|
shadow of prion protein homolog (zebrafish)
|
|
chr12_+_2032649
|
2.514
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr7_+_86111803
|
2.509
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr7_+_86112012
|
2.492
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr11_-_2248757
|
2.480
|
NM_005170
|
ASCL2
|
achaete-scute complex homolog 2 (Drosophila)
|
|
chr16_+_82559600
|
2.475
|
NM_019065
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr13_-_50315832
|
2.450
|
NM_198989
|
DLEU7
|
deleted in lymphocytic leukemia, 7
|
|
chr10_+_133850325
|
2.409
|
NM_006426
|
DPYSL4
|
dihydropyrimidinase-like 4
|
|
chr15_+_77511912
|
2.403
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr14_+_105012106
|
2.397
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr4_-_46690197
|
2.361
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr7_-_158630223
|
2.358
|
NM_003382
|
VIPR2
|
vasoactive intestinal peptide receptor 2
|
|
chr19_+_1397268
|
2.298
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr14_+_73775927
|
2.289
|
NM_182894
|
VSX2
|
visual system homeobox 2
|
|
chr2_-_278181
|
2.267
|
NM_001002919
|
FAM150B
|
family with sequence similarity 150, member B
|
|
chr20_+_20296744
|
2.251
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr22_-_45311730
|
2.249
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr14_+_99181232
|
2.229
|
NM_001127258
NM_032425
|
HHIPL1
|
HHIP-like 1
|
|
chr5_+_170668892
|
2.213
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr1_-_50661716
|
2.211
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr2_-_230287442
|
2.151
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr11_+_627268
|
2.132
|
NM_000797
|
DRD4
|
dopamine receptor D4
|
|
chr20_-_538909
|
2.126
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr19_-_19233370
|
2.093
|
|
HAPLN4
|
hyaluronan and proteoglycan link protein 4
|
|
chr9_+_94987032
|
2.080
|
NM_006648
|
WNK2
|
WNK lysine deficient protein kinase 2
|
|
chr22_+_24895623
|
2.066
|
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr3_-_48607596
|
2.044
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr10_+_69990050
|
2.029
|
NM_030625
|
TET1
|
tet oncogene 1
|
|
chr11_-_64268841
|
2.017
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr1_-_1525309
|
1.997
|
|
LOC643988
|
hypothetical LOC643988
|
|
chr22_+_42138034
|
1.992
|
|
MPPED1
|
metallophosphoesterase domain containing 1
|
|
chr4_+_3737872
|
1.986
|
|
ADRA2C
|
adrenergic, alpha-2C-, receptor
|
|
chr2_+_202607554
|
1.985
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr11_+_110675175
|
1.985
|
NM_001136105
|
C11orf93
|
chromosome 11 open reading frame 93
|
|
chr3_-_10724715
|
1.983
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr18_-_4445247
|
1.972
|
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr5_+_133478252
|
1.957
|
NM_003202
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr10_-_128884411
|
1.956
|
NM_001039762
|
FAM196A
|
family with sequence similarity 196, member A
|
|
chr12_+_130878870
|
1.954
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr1_+_177979037
|
1.953
|
|
FAM163A
|
family with sequence similarity 163, member A
|
|
chr2_+_231610438
|
1.943
|
NM_001144994
|
C2orf72
|
chromosome 2 open reading frame 72
|
|
chr6_+_69401660
|
1.912
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr15_+_90738108
|
1.889
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr4_-_13158391
|
1.884
|
|
LOC285548
|
hypothetical LOC285548
|
|
chr22_+_42137955
|
1.883
|
NM_001044370
|
MPPED1
|
metallophosphoesterase domain containing 1
|
|
chr4_+_3738093
|
1.874
|
NM_000683
|
ADRA2C
|
adrenergic, alpha-2C-, receptor
|
|
chr11_-_64269501
|
1.871
|
NM_153819
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr12_-_56418295
|
1.870
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr12_+_111980044
|
1.870
|
NM_004416
|
DTX1
|
deltex homolog 1 (Drosophila)
|
|
chr22_-_28041474
|
1.866
|
|
RASL10A
|
RAS-like, family 10, member A
|
|
chr16_+_54782802
|
1.821
|
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr3_+_71885890
|
1.807
|
NM_018971
|
GPR27
|
G protein-coupled receptor 27
|
|
chr2_+_100087812
|
1.797
|
|
|
|
|
chr19_+_38377329
|
1.795
|
NM_002333
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr2_-_204108151
|
1.792
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr1_-_925332
|
1.786
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr1_-_26245031
|
1.763
|
NM_001004434
NM_032513
|
SLC30A2
|
solute carrier family 30 (zinc transporter), member 2
|
|
chr11_+_359723
|
1.745
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr22_+_24895454
|
1.742
|
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr8_+_26427378
|
1.737
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr1_+_52871603
|
1.718
|
NM_001042693
|
FAM159A
|
family with sequence similarity 159, member A
|
|
chr22_-_43783449
|
1.702
|
|
PHF21B
|
PHD finger protein 21B
|
|
chr22_+_24895424
|
1.695
|
NM_001184773
NM_001184774
NM_001184775
NM_001184776
NM_001184777
NM_021115
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr17_-_29930431
|
1.686
|
NM_207454
|
C17orf102
|
chromosome 17 open reading frame 102
|
|
chrX_-_71442488
|
1.681
|
NM_001144885
NM_001144887
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1
|
|
chr11_+_17697685
|
1.681
|
NM_002478
|
MYOD1
|
myogenic differentiation 1
|
|
chr4_+_4912272
|
1.675
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chrX_+_152561086
|
1.675
|
NM_001395
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr16_-_695719
|
1.671
|
NM_153350
|
FBXL16
|
F-box and leucine-rich repeat protein 16
|
|
chr3_+_3816079
|
1.670
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr6_+_137285071
|
1.669
|
NM_001008783
|
SLC35D3
|
solute carrier family 35, member D3
|
|
chr11_+_14621972
|
1.665
|
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr12_+_5411534
|
1.656
|
NM_001102654
|
NTF3
|
neurotrophin 3
|
|
chr10_+_94439658
|
1.645
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chrX_-_132376660
|
1.641
|
NM_001448
|
GPC4
|
glypican 4
|
|
chr1_-_23623713
|
1.625
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr3_-_69064364
|
1.622
|
NM_001005527
NM_182522
|
FAM19A4
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4
|
|
chr22_+_49386044
|
1.622
|
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr1_+_235272324
|
1.612
|
NM_001035
|
RYR2
|
ryanodine receptor 2 (cardiac)
|
|
chr17_-_34015631
|
1.607
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr14_-_99695684
|
1.599
|
NM_206918
|
DEGS2
|
degenerative spermatocyte homolog 2, lipid desaturase (Drosophila)
|
|
chr2_-_128792515
|
1.592
|
NM_004807
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chr14_+_95412481
|
1.589
|
|
|
|
|
chr12_+_48641545
|
1.579
|
NM_001651
|
AQP5
|
aquaporin 5
|
|
chr1_+_2975590
|
1.577
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr13_+_48692472
|
1.576
|
NM_001507
|
MLNR
|
motilin receptor
|
|
chr15_+_78483624
|
1.572
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr7_+_3307445
|
1.562
|
NM_152744
|
SDK1
|
sidekick homolog 1, cell adhesion molecule (chicken)
|
|
chr5_+_38294289
|
1.562
|
NM_152403
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr22_-_43784093
|
1.560
|
NM_138415
|
PHF21B
|
PHD finger protein 21B
|
|
chr11_-_69343128
|
1.559
|
NM_005247
|
FGF3
|
fibroblast growth factor 3
|
|
chr1_+_226937732
|
1.557
|
|
RHOU
|
ras homolog gene family, member U
|
|
chr10_+_134751398
|
1.552
|
NM_001083909
|
GPR123
|
G protein-coupled receptor 123
|
|
chr21_+_42946719
|
1.548
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr20_-_22514092
|
1.544
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr17_-_5344898
|
1.540
|
NM_001162371
|
LOC728392
|
hypothetical protein LOC728392
|
|
chr9_+_78824390
|
1.537
|
NM_001013735
|
FOXB2
|
forkhead box B2
|
|
chr12_+_131705438
|
1.531
|
NM_012226
NM_016318
NM_170682
NM_170683
NM_174872
NM_174873
|
P2RX2
|
purinergic receptor P2X, ligand-gated ion channel, 2
|
|
chr4_+_2031036
|
1.525
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr4_+_36923084
|
1.520
|
NM_001144990
|
KIAA1239
|
KIAA1239
|
|
chr10_+_50488352
|
1.514
|
NM_003055
|
SLC18A3
|
solute carrier family 18 (vesicular acetylcholine), member 3
|
|
chr10_+_23521464
|
1.506
|
NM_178161
|
PTF1A
|
pancreas specific transcription factor, 1a
|
|
chr22_+_47263919
|
1.505
|
NM_001082967
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr16_+_25610847
|
1.498
|
NM_006040
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4
|
|
chr16_+_1143241
|
1.497
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr20_-_60484420
|
1.489
|
NM_080473
|
GATA5
|
GATA binding protein 5
|
|
chr1_-_38285033
|
1.485
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr2_+_5750229
|
1.482
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr1_-_925215
|
1.481
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr16_+_65754788
|
1.470
|
NM_001040667
NM_001538
|
HSF4
|
heat shock transcription factor 4
|
|
chr6_-_165996031
|
1.468
|
|
LOC100132188
|
LP7097
|
|
chr1_-_239587007
|
1.455
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr2_+_936534
|
1.453
|
NM_018968
|
SNTG2
|
syntrophin, gamma 2
|
|
chr22_+_28446288
|
1.446
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr6_+_159510416
|
1.439
|
NM_032532
|
FNDC1
|
fibronectin type III domain containing 1
|
|
chr17_+_11085464
|
1.436
|
NM_001173461
NM_001173462
NM_207386
|
SHISA6
|
shisa homolog 6 (Xenopus laevis)
|
|
chr9_+_70509925
|
1.434
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr19_-_16998449
|
1.431
|
NM_015692
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8
|
|
chr16_-_2186465
|
1.430
|
NM_020764
|
CASKIN1
|
CASK interacting protein 1
|
|
chr13_+_32488477
|
1.424
|
NM_004795
|
KL
|
klotho
|
|
chr10_-_88116181
|
1.423
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chrX_-_138114100
|
1.422
|
|
FGF13
|
fibroblast growth factor 13
|
|
chr17_-_47590975
|
1.421
|
|
CA10
|
carbonic anhydrase X
|
|
chr18_-_4445265
|
1.420
|
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr8_-_26427304
|
1.418
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr22_+_27931685
|
1.409
|
NM_133455
|
EMID1
|
EMI domain containing 1
|
|
chr2_-_106869488
|
1.407
|
|
|
|
|
chr5_+_149526533
|
1.400
|
NM_001804
|
CDX1
|
caudal type homeobox 1
|
|
chr16_-_49742651
|
1.393
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr16_+_88169612
|
1.389
|
NM_014427
NM_153636
|
CPNE7
|
copine VII
|
|
chr19_-_18198234
|
1.387
|
NM_001098818
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific
|
|
chr17_-_47591123
|
1.387
|
NM_020178
|
CA10
|
carbonic anhydrase X
|
|
chr8_+_136538739
|
1.376
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr1_-_57661373
|
1.366
|
|
DAB1
|
disabled homolog 1 (Drosophila)
|
|
chr16_-_30288988
|
1.365
|
NM_015527
|
TBC1D10B
|
TBC1 domain family, member 10B
|
|
chr8_-_133562185
|
1.362
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr3_+_13565644
|
1.361
|
|
FBLN2
|
fibulin 2
|
|
chr3_-_50515857
|
1.361
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr1_-_218168548
|
1.361
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chrX_-_8660132
|
1.360
|
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chr19_+_659766
|
1.355
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr9_+_99656013
|
1.349
|
|
|
|
|
chr14_-_59113301
|
1.346
|
NM_001164399
|
C14orf38
|
chromosome 14 open reading frame 38
|
|
chr12_+_2774362
|
1.345
|
NM_002014
|
FKBP4
|
FK506 binding protein 4, 59kDa
|
|
chr5_+_3649167
|
1.335
|
NM_024337
|
IRX1
|
iroquois homeobox 1
|
|
chr22_-_22511173
|
1.335
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chr17_+_40089270
|
1.332
|
NM_001145080
|
C17orf104
|
chromosome 17 open reading frame 104
|
|
chr7_+_100156358
|
1.324
|
NM_000799
|
EPO
|
erythropoietin
|
|
chr17_+_27617307
|
1.324
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr2_+_208979800
|
1.322
|
NM_005048
|
PTH2R
|
parathyroid hormone 2 receptor
|
|
chr22_-_37180958
|
1.321
|
NM_152868
|
KCNJ4
|
potassium inwardly-rectifying channel, subfamily J, member 4
|
|
chr4_+_77391876
|
1.319
|
NM_001136570
|
FAM47E
|
family with sequence similarity 47, member E
|
|
chr3_-_129694717
|
1.315
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr10_+_50487146
|
1.309
|
NM_020984
|
CHAT
|
choline O-acetyltransferase
|
|
chr1_+_226937424
|
1.309
|
NM_021205
|
RHOU
|
ras homolog gene family, member U
|
|
chr17_+_56831951
|
1.302
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr11_-_27678551
|
1.298
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_-_21818881
|
1.294
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr11_-_114880275
|
1.292
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|