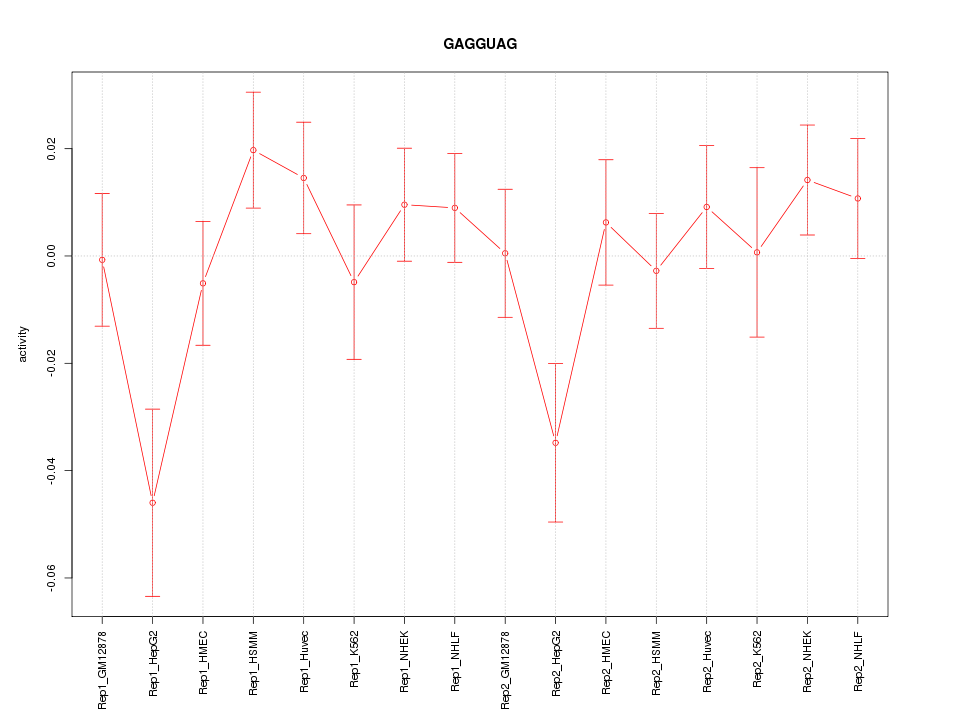
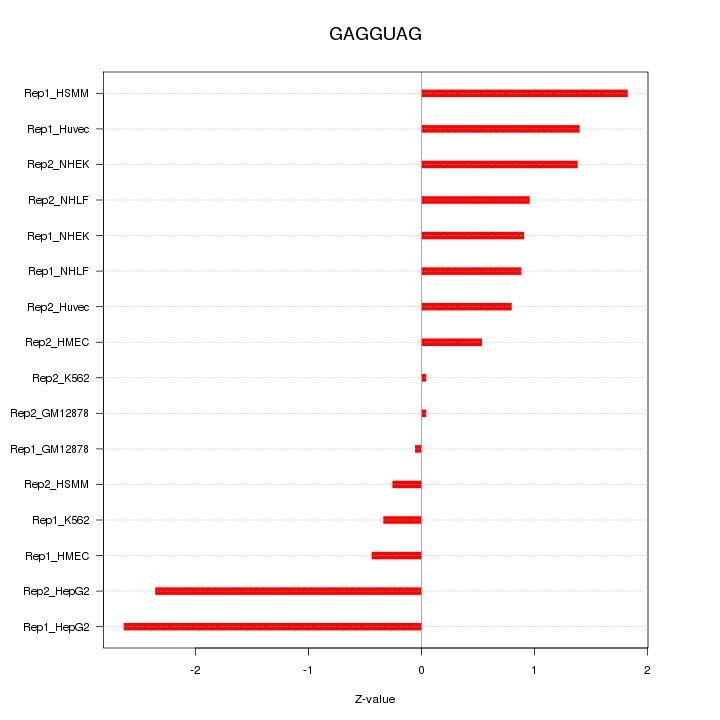
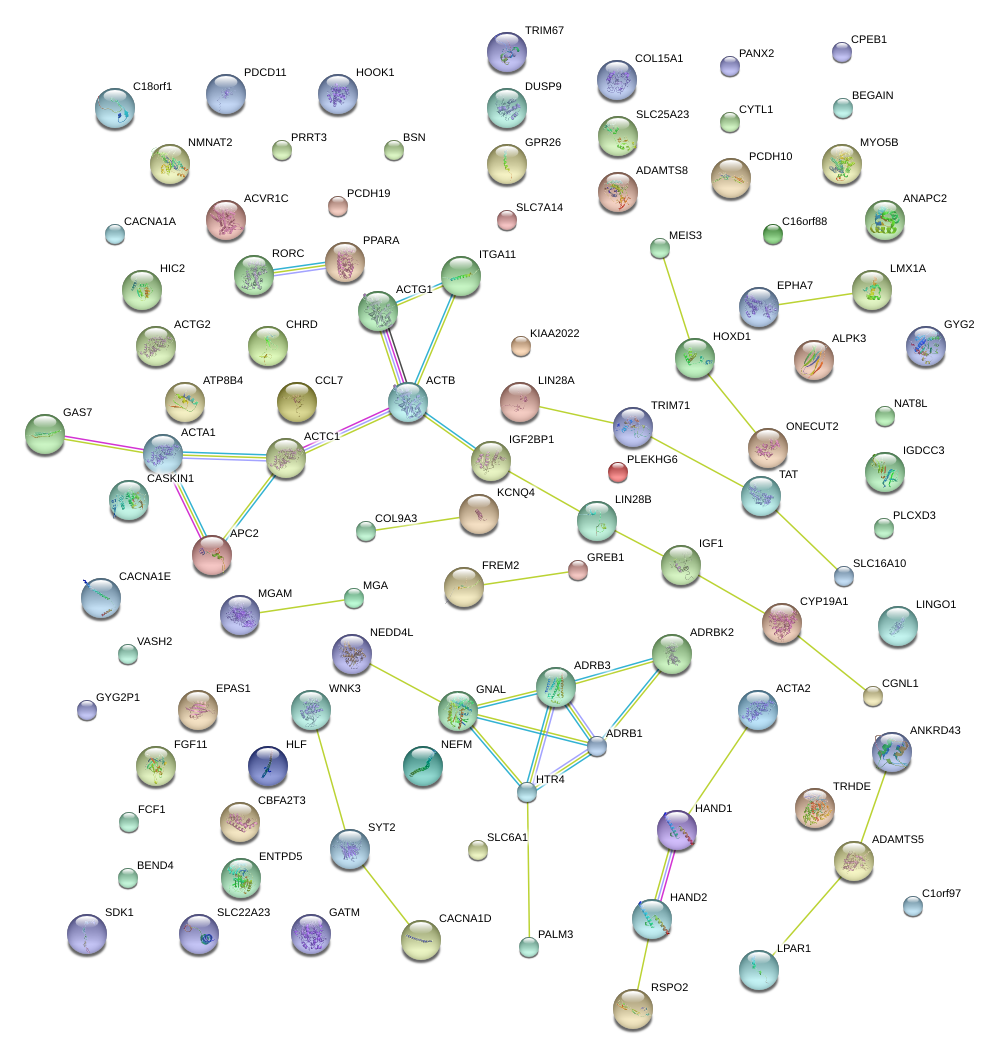
|
chr5_+_132176921
|
2.152
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr9_+_100745531
|
2.138
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr15_-_63457378
|
2.014
|
NM_004884
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3
|
|
chr18_+_53253775
|
2.013
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr3_-_171786468
|
1.979
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr3_+_32834513
|
1.940
|
NM_001039111
|
TRIM71
|
tripartite motif containing 71
|
|
chr8_-_37943143
|
1.933
|
NM_000025
|
ADRB3
|
adrenergic, beta-3-, receptor
|
|
chr2_-_158162271
|
1.900
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr19_-_14030970
|
1.898
|
NM_001145028
|
PALM3
|
paralemmin 3
|
|
chr4_-_41849651
|
1.872
|
NM_001159547
NM_207406
|
BEND4
|
BEN domain containing 4
|
|
chr15_-_81113676
|
1.827
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr17_+_50697319
|
1.643
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr17_+_44429748
|
1.638
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr20_+_60918836
|
1.611
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr1_-_163592098
|
1.568
|
NM_001174069
|
LMX1A
|
LIM homeobox transcription factor 1, alpha
|
|
chr16_-_87535106
|
1.560
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr18_+_13601664
|
1.557
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr8_+_24828359
|
1.525
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr1_-_163592575
|
1.507
|
NM_177398
|
LMX1A
|
LIM homeobox transcription factor 1, alpha
|
|
chr15_-_43458247
|
1.458
|
NM_001482
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr18_-_45975163
|
1.434
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr18_+_13208777
|
1.434
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr1_+_229365296
|
1.422
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chrX_+_152561086
|
1.409
|
NM_001395
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr1_-_150065176
|
1.405
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr6_+_111515473
|
1.342
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr17_-_10042579
|
1.340
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr17_+_7283412
|
1.327
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr8_+_24827173
|
1.319
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr1_+_41022066
|
1.315
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr10_+_115793795
|
1.274
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr15_+_39739864
|
1.270
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr18_+_11741470
|
1.258
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr5_-_148013908
|
1.238
|
NM_000870
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr3_+_11009419
|
1.227
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr1_+_60053050
|
1.218
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr14_-_100104014
|
1.213
|
NM_001159531
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chrX_+_2756810
|
1.213
|
NM_001079855
NM_001184702
NM_001184703
NM_001184704
NM_003918
|
GYG2
|
glycogenin 2
|
|
chr1_+_211190476
|
1.187
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr8_-_109165012
|
1.164
|
NM_178565
|
RSPO2
|
R-spondin 2 homolog (Xenopus laevis)
|
|
chr1_-_200946038
|
1.163
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr6_+_105511615
|
1.153
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr1_-_227636462
|
1.152
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr3_+_185580554
|
1.137
|
NM_003741
|
CHRD
|
chordin
|
|
chr16_-_2186465
|
1.133
|
NM_020764
|
CASKIN1
|
CASK interacting protein 1
|
|
chr22_+_24290738
|
1.129
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr15_+_83160914
|
1.126
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr12_-_101396459
|
1.107
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr5_-_41546384
|
1.098
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr16_-_70168449
|
1.078
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr2_+_176761552
|
1.075
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr17_+_29621352
|
1.063
|
NM_006273
|
CCL7
|
chemokine (C-C motif) ligand 7
|
|
chr4_+_2031036
|
1.046
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chrX_-_99551926
|
1.036
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr15_-_49418027
|
1.032
|
NM_000103
NM_031226
|
CYP19A1
|
cytochrome P450, family 19, subfamily A, polypeptide 1
|
|
chr21_-_27261276
|
1.027
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr15_+_55455994
|
1.025
|
NM_032866
|
CGNL1
|
cingulin-like 1
|
|
chr22_+_44925162
|
1.013
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr13_+_38159172
|
1.010
|
NM_207361
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr15_-_75711743
|
1.010
|
NM_032808
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr1_+_26609819
|
1.001
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr7_+_3307445
|
1.000
|
NM_152744
|
SDK1
|
sidekick homolog 1, cell adhesion molecule (chicken)
|
|
chrX_-_74062005
|
0.995
|
NM_001008537
|
KIAA2022
|
KIAA2022
|
|
chr19_-_52614550
|
0.991
|
NM_001009813
NM_020160
|
MEIS3
|
Meis homeobox 3
|
|
chr5_-_153838013
|
0.982
|
NM_004821
|
HAND1
|
heart and neural crest derivatives expressed 1
|
|
chr6_-_94185780
|
0.977
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr1_+_179719338
|
0.976
|
NM_000721
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr12_+_6292077
|
0.965
|
NM_001144857
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr22_+_20101661
|
0.959
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr15_-_48198697
|
0.955
|
NM_024837
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr2_+_11591692
|
0.953
|
NM_014668
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr18_+_54013575
|
0.951
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr15_-_66511488
|
0.935
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr4_-_174687931
|
0.929
|
NM_021973
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr22_+_48951286
|
0.928
|
NM_001160300
NM_052839
|
PANX2
|
pannexin 2
|
|
chr19_+_1401117
|
0.927
|
NM_005883
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr11_-_129803712
|
0.915
|
NM_007037
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr1_-_181654124
|
0.895
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr3_+_53504070
|
0.886
|
NM_000720
NM_001128839
NM_001128840
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr12_+_70952729
|
0.857
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr6_-_3390197
|
0.855
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr3_+_49566901
|
0.853
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr10_+_125415860
|
0.850
|
NM_153442
|
GPR26
|
G protein-coupled receptor 26
|
|
chrX_-_54401161
|
0.846
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr12_+_56435042
|
0.846
|
NM_138396
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9
|
|
chr3_-_130807865
|
0.844
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr5_-_35266550
|
0.844
|
NM_000949
|
PRLR
|
prolactin receptor
|
|
chr6_-_90178685
|
0.842
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr1_-_24386324
|
0.842
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr12_+_77781903
|
0.841
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr6_+_87703742
|
0.840
|
NM_000865
|
HTR1E
|
5-hydroxytryptamine (serotonin) receptor 1E
|
|
chr19_-_52667056
|
0.831
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr1_-_115682374
|
0.825
|
NM_002506
|
NGF
|
nerve growth factor (beta polypeptide)
|
|
chr19_+_51492136
|
0.815
|
NM_152795
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr18_+_53967544
|
0.804
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr5_+_176170041
|
0.799
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr1_+_29435534
|
0.780
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr1_-_177378801
|
0.778
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr9_-_10602722
|
0.765
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr3_+_103636548
|
0.762
|
NM_175056
|
ZPLD1
|
zona pellucida-like domain containing 1
|
|
chr22_+_44083485
|
0.761
|
NM_001104595
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr9_-_100510657
|
0.754
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr15_-_36644122
|
0.743
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr3_-_53055078
|
0.739
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr14_-_64508547
|
0.735
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr19_-_7244876
|
0.731
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr17_-_9803490
|
0.731
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr1_-_45444812
|
0.729
|
NM_020883
|
ZSWIM5
|
zinc finger, SWIM-type containing 5
|
|
chr1_-_180840016
|
0.708
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr5_+_149131695
|
0.707
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chrX_+_72699686
|
0.701
|
NM_001039840
|
CHIC1
|
cysteine-rich hydrophobic domain 1
|
|
chr11_-_117521344
|
0.701
|
NM_001142349
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chrX_+_147608302
|
0.696
|
NM_001170628
|
AFF2
|
AF4/FMR2 family, member 2
|
|
chr16_+_49139694
|
0.692
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr1_-_202647526
|
0.687
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory (inhibitor) subunit 15B
|
|
chr13_-_51483473
|
0.687
|
NM_000053
NM_001005918
|
ATP7B
|
ATPase, Cu++ transporting, beta polypeptide
|
|
chr12_-_50502474
|
0.686
|
NM_001013690
|
FIGNL2
|
fidgetin-like 2
|
|
chrX_+_104953187
|
0.686
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr19_+_5774817
|
0.685
|
NM_004558
|
NRTN
|
neurturin
|
|
chr15_+_49831030
|
0.681
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr6_+_101953581
|
0.679
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr21_-_44955889
|
0.678
|
NM_144991
|
TSPEAR
|
thrombospondin-type laminin G domain and EAR repeats
|
|
chr9_-_103540682
|
0.678
|
NM_133445
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A
|
|
chr12_-_10215942
|
0.672
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr7_-_23476497
|
0.672
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr21_-_41801778
|
0.671
|
NM_001135099
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
chr7_+_100584397
|
0.671
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr1_+_203804734
|
0.669
|
NM_181644
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr7_-_71440143
|
0.662
|
NM_001017440
|
CALN1
|
calneuron 1
|
|
chr17_+_26742767
|
0.661
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr9_-_76692082
|
0.658
|
NM_001177311
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|
|
chr13_-_32678142
|
0.655
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_-_200879203
|
0.644
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr9_+_135314907
|
0.641
|
NM_001135775
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr8_-_13018123
|
0.638
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr5_+_56146656
|
0.631
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr1_-_86394695
|
0.630
|
NM_152890
|
COL24A1
|
collagen, type XXIV, alpha 1
|
|
chr22_-_37966852
|
0.628
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog)
|
|
chr18_+_13610866
|
0.628
|
NM_004338
NM_181483
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr11_-_133786935
|
0.627
|
NM_018644
NM_054025
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P)
|
|
chr1_-_40903897
|
0.626
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr12_+_4253143
|
0.623
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr19_+_51493486
|
0.621
|
NM_022462
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chrX_-_79951838
|
0.621
|
NM_153252
|
BRWD3
|
bromodomain and WD repeat domain containing 3
|
|
chr12_+_4788602
|
0.617
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr15_-_27650218
|
0.616
|
NM_015307
|
FAM189A1
|
family with sequence similarity 189, member A1
|
|
chr17_+_31082791
|
0.616
|
NM_033315
|
RASL10B
|
RAS-like, family 10, member B
|
|
chr10_+_35575958
|
0.615
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr22_-_21193504
|
0.613
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr15_-_63502438
|
0.611
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr11_+_45825244
|
0.608
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr3_-_121445831
|
0.606
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr7_-_6557591
|
0.603
|
NM_001145118
|
GRID2IP
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein
|
|
chr7_+_140420500
|
0.603
|
NM_001195278
|
LOC100131199
|
transmembrane protein 178-like
|
|
chr22_-_37970902
|
0.602
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog)
|
|
chr15_+_38520594
|
0.598
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr9_+_4652285
|
0.596
|
NM_203453
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2
|
|
chr2_-_96899396
|
0.594
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr22_+_36422940
|
0.592
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr21_+_26029128
|
0.590
|
NM_001197297
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr2_+_204509698
|
0.587
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr18_-_43711452
|
0.584
|
NM_001003652
NM_001135937
|
SMAD2
|
SMAD family member 2
|
|
chr1_+_158636987
|
0.583
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr5_+_173247919
|
0.577
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr8_-_22606653
|
0.576
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr12_+_93066370
|
0.573
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr12_-_113330574
|
0.572
|
NM_000192
NM_080717
|
TBX5
|
T-box 5
|
|
chr17_-_7138590
|
0.564
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr21_-_41801950
|
0.562
|
NM_005656
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
chr10_+_102722275
|
0.561
|
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr9_-_98368913
|
0.559
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr15_+_40484249
|
0.558
|
NM_173089
NM_173090
|
CAPN3
|
calpain 3, (p94)
|
|
chr8_-_70909754
|
0.555
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr16_-_27982277
|
0.554
|
NM_001109763
|
GSG1L
|
GSG1-like
|
|
chr12_-_47735373
|
0.553
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr4_-_1156959
|
0.551
|
NM_001128325
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr4_+_76077229
|
0.551
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr9_+_89302574
|
0.550
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chrX_+_118254233
|
0.549
|
NM_006667
|
PGRMC1
|
progesterone receptor membrane component 1
|
|
chr8_-_17624009
|
0.541
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr16_+_28211123
|
0.540
|
NM_001024401
|
SBK1
|
SH3-binding domain kinase 1
|
|
chr22_+_38296703
|
0.539
|
NM_001003406
NM_021096
|
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit
|
|
chrX_-_111953009
|
0.536
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr17_+_8865547
|
0.535
|
NM_004822
|
NTN1
|
netrin 1
|
|
chrX_-_24575375
|
0.534
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr13_-_32658215
|
0.531
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr12_-_47679176
|
0.531
|
NM_015086
|
DDN
|
dendrin
|
|
chr16_-_2848106
|
0.528
|
NM_022119
|
PRSS22
|
protease, serine, 22
|
|
chr12_-_24946495
|
0.523
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chrX_-_20194629
|
0.522
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr11_+_17714070
|
0.522
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr4_-_171161486
|
0.521
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr15_+_40481879
|
0.518
|
NM_173088
|
CAPN3
|
calpain 3, (p94)
|
|
chr21_+_36993860
|
0.516
|
NM_005069
NM_009586
|
SIM2
|
single-minded homolog 2 (Drosophila)
|
|
chr1_+_32417909
|
0.514
|
NM_175852
|
TXLNA
|
taxilin alpha
|
|
chr5_-_131375148
|
0.508
|
NM_001009185
NM_015256
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr1_-_202387767
|
0.507
|
NM_018208
|
ETNK2
|
ethanolamine kinase 2
|
|
chr14_-_100105883
|
0.504
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr22_+_22445035
|
0.503
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chrX_-_15263596
|
0.501
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|