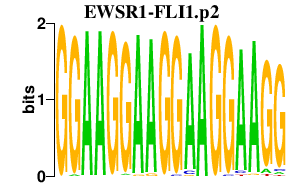
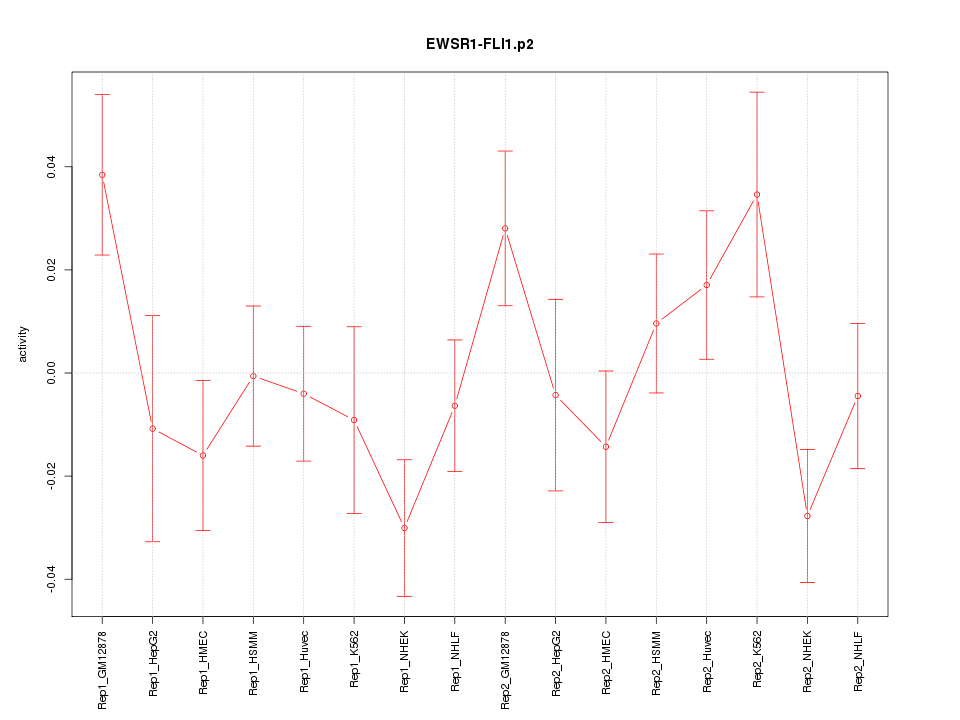
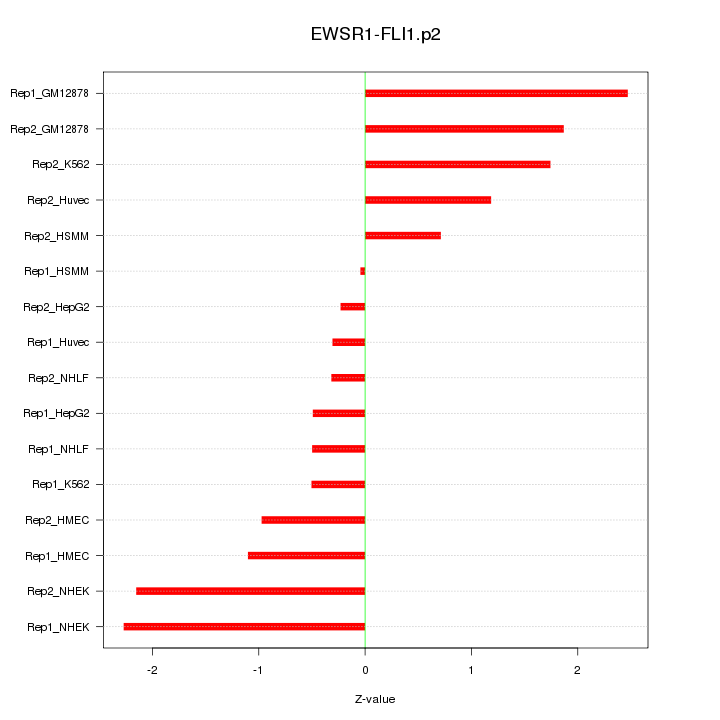
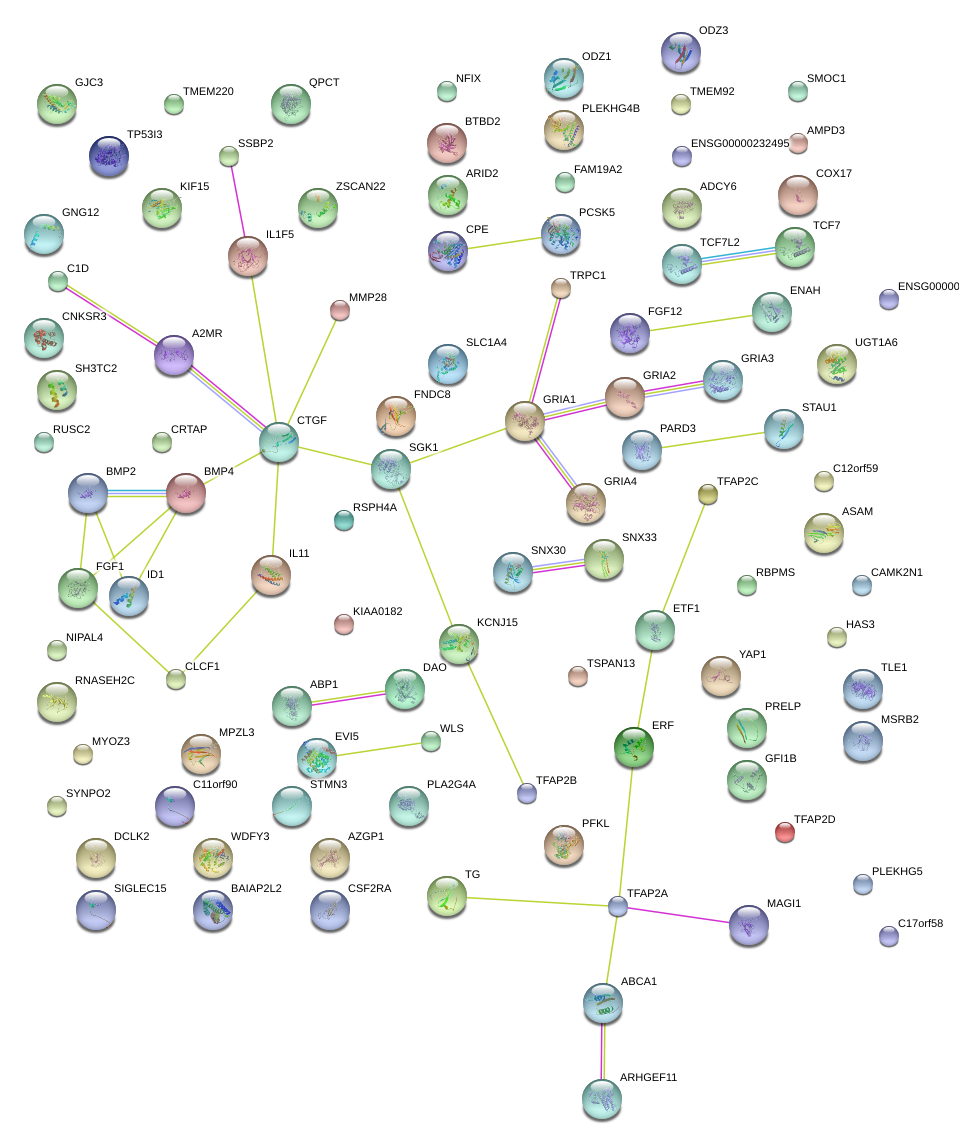
|
chr4_+_120029426
|
2.588
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr17_-_31146660
|
2.215
|
NM_001032278
NM_024302
|
MMP28
|
matrix metallopeptidase 28
|
|
chr14_-_53493198
|
2.126
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr6_-_10520592
|
1.954
|
NM_001032280
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr4_-_86106567
|
1.953
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chrX_+_1361570
|
1.900
|
NM_001161530
NM_172247
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr11_+_101485946
|
1.855
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr1_-_223907283
|
1.796
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr6_-_10520264
|
1.781
|
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr2_+_65069081
|
1.702
|
NM_001193493
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr2_+_113533155
|
1.656
|
NM_012275
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr2_+_113532685
|
1.631
|
NM_173170
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr1_-_68470585
|
1.481
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr1_-_68071635
|
1.422
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr2_+_37425240
|
1.398
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr1_-_6468600
|
1.192
|
NM_001042665
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr4_+_183302105
|
1.178
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr1_-_93030538
|
1.162
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr19_-_1966628
|
1.159
|
NM_017797
|
BTBD2
|
BTB (POZ) domain containing 2
|
|
chr6_-_132314004
|
1.113
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr4_+_166519392
|
1.092
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr9_+_134843918
|
1.069
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr7_+_16759845
|
1.041
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr5_+_156819782
|
1.036
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr9_+_77695334
|
1.024
|
NM_001190482
NM_006200
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
|
chr10_+_114700775
|
1.010
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr12_+_10222823
|
1.008
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr11_-_122571198
|
1.007
|
NM_024769
|
CLMP
|
CXADR-like membrane protein
|
|
chr19_+_12996830
|
1.007
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr3_-_65999548
|
1.002
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr12_+_55808696
|
0.968
|
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr16_+_67697622
|
0.963
|
NM_138612
|
HAS3
|
hyaluronan synthase 3
|
|
chr6_-_134538762
|
0.852
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_+_55808514
|
0.835
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr8_+_30361485
|
0.831
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr9_+_114552871
|
0.773
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr18_+_41659542
|
0.752
|
NM_213602
|
SIGLEC15
|
sialic acid binding Ig-like lectin 15
|
|
chr5_+_156819604
|
0.750
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr1_+_201711578
|
0.728
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr7_-_99365178
|
0.718
|
NM_181538
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa
|
|
chr19_+_12996745
|
0.712
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr10_-_35143900
|
0.693
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr7_-_99411573
|
0.680
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr11_-_122571046
|
0.640
|
|
CLMP
|
CXADR-like membrane protein
|
|
chr17_+_45706786
|
0.619
|
NM_153229
|
TMEM92
|
transmembrane protein 92
|
|
chr1_-_72521079
|
0.607
|
|
|
|
|
chr9_+_35528890
|
0.590
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr5_-_148422799
|
0.587
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr12_+_107797985
|
0.568
|
NM_001917
|
DAO
|
D-amino-acid oxidase
|
|
chr8_+_133948384
|
0.559
|
NM_003235
|
TG
|
thyroglobulin
|
|
chr3_-_120878910
|
0.541
|
NM_005694
|
COX17
|
COX17 cytochrome c oxidase assembly homolog (S. cerevisiae)
|
|
chr11_-_66897584
|
0.534
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr11_-_117628211
|
0.526
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr1_+_185064654
|
0.501
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr9_-_83494198
|
0.497
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr3_+_143925904
|
0.495
|
NM_003304
|
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1
|
|
chr7_+_21434818
|
0.491
|
|
|
|
|
chr21_+_44544381
|
0.489
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr19_-_47451119
|
0.477
|
NM_006494
|
ERF
|
Ets2 repressor factor
|
|
chr5_-_81082699
|
0.471
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr20_-_61755145
|
0.471
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr21_+_44544392
|
0.470
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr12_-_47464143
|
0.469
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chr20_+_6696744
|
0.460
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr21_+_44544349
|
0.459
|
NM_002626
|
PFKL
|
phosphofructokinase, liver
|
|
chr10_+_23424383
|
0.458
|
NM_012228
|
MSRB2
|
methionine sulfoxide reductase B2
|
|
chr16_+_84202515
|
0.458
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr6_-_134538526
|
0.455
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_-_155281785
|
0.454
|
NM_014784
NM_198236
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11
|
|
chr17_-_10574370
|
0.443
|
NM_001004313
|
TMEM220
|
transmembrane protein 220
|
|
chr3_-_193928038
|
0.434
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr5_+_152850276
|
0.430
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr20_-_47238310
|
0.430
|
NM_001037328
NM_004602
NM_017452
NM_017453
NM_017454
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr5_-_141981083
|
0.425
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr1_-_20685314
|
0.425
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr12_+_44409743
|
0.416
|
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr2_+_96269703
|
0.411
|
NM_001037228
|
LOC285033
|
hypothetical protein LOC285033
|
|
chr5_+_150020595
|
0.409
|
NM_001122853
NM_133371
|
MYOZ3
|
myozenin 3
|
|
chr11_-_65244718
|
0.407
|
NM_032193
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr17_+_30472710
|
0.406
|
NM_017559
|
FNDC8
|
fibronectin type III domain containing 8
|
|
chr19_+_13066956
|
0.405
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_-_92489652
|
0.403
|
|
LOC144481
|
hypothetical protein LOC144481
|
|
chr3_+_33130563
|
0.390
|
|
CRTAP
|
cartilage associated protein
|
|
chr4_+_151218862
|
0.390
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr19_-_60573552
|
0.379
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr11_+_10480891
|
0.375
|
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr6_+_139055244
|
0.375
|
|
FLJ46906
|
hypothetical LOC441172
|
|
chr11_+_10433241
|
0.373
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr11_-_65244486
|
0.370
|
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr11_-_93223315
|
0.367
|
NM_001144871
|
C11orf90
|
chromosome 11 open reading frame 90
|
|
chr20_+_29656744
|
0.367
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr14_+_69415880
|
0.365
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr2_-_24161033
|
0.361
|
NM_147184
NM_004881
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr9_-_106705867
|
0.357
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr6_+_117044334
|
0.349
|
NM_001010892
NM_001161664
|
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas)
|
|
chr21_+_38550533
|
0.347
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr17_-_36346240
|
0.341
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr16_-_2848106
|
0.336
|
NM_022119
|
PRSS22
|
protease, serine, 22
|
|
chr9_-_129679760
|
0.332
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr18_+_44319365
|
0.331
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr3_+_33130451
|
0.329
|
NM_006371
|
CRTAP
|
cartilage associated protein
|
|
chr1_-_231498081
|
0.329
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr1_+_109036431
|
0.328
|
NM_018061
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr2_-_25418249
|
0.328
|
NM_022552
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr8_-_95556460
|
0.321
|
NM_012415
|
RAD54B
|
RAD54 homolog B (S. cerevisiae)
|
|
chr7_-_127458231
|
0.317
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr3_-_188936941
|
0.317
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr18_+_54682261
|
0.308
|
|
ZNF532
|
zinc finger protein 532
|
|
chr1_+_201711505
|
0.307
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr11_+_10434056
|
0.305
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr17_+_43082302
|
0.303
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr9_-_106705780
|
0.300
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr4_+_154345039
|
0.299
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr7_-_106991941
|
0.288
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr2_-_55349818
|
0.288
|
NM_001005369
|
MTIF2
|
mitochondrial translational initiation factor 2
|
|
chr1_-_155936904
|
0.288
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
chr9_-_103397023
|
0.283
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr12_+_44409762
|
0.283
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr19_-_44060729
|
0.282
|
NM_001195833
NM_198445
|
RINL
|
Ras and Rab interactor-like
|
|
chr19_-_60783996
|
0.282
|
NM_152600
|
ZNF579
|
zinc finger protein 579
|
|
chr20_+_43691797
|
0.281
|
NM_080753
|
WFDC10A
|
WAP four-disulfide core domain 10A
|
|
chr6_-_143307976
|
0.276
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr17_+_45488356
|
0.276
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr12_-_13139848
|
0.272
|
NM_031289
NM_153823
|
GSG1
|
germ cell associated 1
|
|
chr10_-_103864579
|
0.266
|
|
LDB1
|
LIM domain binding 1
|
|
chr12_+_54947293
|
0.265
|
NM_001099337
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr13_-_32678142
|
0.264
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr15_-_77085056
|
0.263
|
NM_153815
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr1_+_151596953
|
0.260
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr2_-_55349851
|
0.259
|
NM_002453
|
MTIF2
|
mitochondrial translational initiation factor 2
|
|
chr1_+_26895108
|
0.257
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr22_+_38296703
|
0.257
|
NM_001003406
NM_021096
|
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit
|
|
chr7_+_36395913
|
0.252
|
NM_018685
|
ANLN
|
anillin, actin binding protein
|
|
chr11_-_66898223
|
0.245
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr12_+_92295738
|
0.241
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr14_-_20636568
|
0.238
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr1_-_202730135
|
0.235
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr7_+_116182676
|
0.234
|
|
|
|
|
chr7_-_99604127
|
0.232
|
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr8_-_145530639
|
0.231
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr12_+_14409875
|
0.227
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr2_+_149118791
|
0.226
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr12_+_54947180
|
0.226
|
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr15_+_39310757
|
0.224
|
|
CHP
|
calcium binding protein P22
|
|
chr15_+_88345612
|
0.223
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr7_+_143043871
|
0.223
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr19_-_47450991
|
0.222
|
|
ERF
|
Ets2 repressor factor
|
|
chr4_+_84676280
|
0.222
|
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr10_+_106004457
|
0.221
|
NM_001191002
NM_004832
|
GSTO1
|
glutathione S-transferase omega 1
|
|
chr1_-_26105477
|
0.220
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr1_+_44185005
|
0.219
|
NM_014652
|
IPO13
|
importin 13
|
|
chr1_-_20684858
|
0.216
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr12_-_54869511
|
0.213
|
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr5_+_86599350
|
0.213
|
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr12_-_48017223
|
0.212
|
NM_001008223
|
C1QL4
|
complement component 1, q subcomponent-like 4
|
|
chr9_+_35528628
|
0.203
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr2_+_182464910
|
0.203
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr1_+_93317374
|
0.202
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr20_-_48741481
|
0.198
|
|
|
|
|
chr5_-_81082614
|
0.197
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr1_-_224563820
|
0.196
|
NM_173083
|
LIN9
|
lin-9 homolog (C. elegans)
|
|
chr11_+_73034870
|
0.196
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr3_+_143925545
|
0.193
|
|
|
|
|
chr11_-_76800525
|
0.192
|
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr10_+_75211810
|
0.192
|
NM_203298
|
CHCHD1
|
coiled-coil-helix-coiled-coil-helix domain containing 1
|
|
chrX_+_67635348
|
0.190
|
NM_001195214
NM_173834
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr1_-_72520735
|
0.189
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr1_-_180297372
|
0.189
|
NM_001009992
|
ZNF648
|
zinc finger protein 648
|
|
chr17_-_34157962
|
0.188
|
NM_007144
|
PCGF2
|
polycomb group ring finger 2
|
|
chr7_+_128142661
|
0.188
|
NM_032599
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr8_-_22982435
|
0.188
|
NM_003842
NM_147187
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr2_+_79593567
|
0.187
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr16_-_69277194
|
0.187
|
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr1_-_224564038
|
0.186
|
|
LIN9
|
lin-9 homolog (C. elegans)
|
|
chr17_+_45488330
|
0.186
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr2_+_182464713
|
0.185
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chr21_+_38550739
|
0.180
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr11_-_8788765
|
0.179
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr5_-_38631263
|
0.177
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr7_-_99604171
|
0.175
|
NM_024637
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr1_+_109969571
|
0.175
|
NM_203404
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr1_-_171442937
|
0.174
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr15_-_56829225
|
0.168
|
NM_001110
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr8_-_38127489
|
0.167
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chr16_-_69277355
|
0.167
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr17_-_34607257
|
0.166
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr19_+_54786723
|
0.166
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr11_-_8789457
|
0.166
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr3_-_197529434
|
0.164
|
NM_152773
|
TCTEX1D2
|
Tctex1 domain containing 2
|
|
chr16_-_79811475
|
0.164
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr17_-_8004671
|
0.163
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr19_-_53515143
|
0.163
|
NM_144577
|
CCDC114
|
coiled-coil domain containing 114
|
|
chr16_-_30841938
|
0.161
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr3_+_188131209
|
0.159
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr19_-_14447122
|
0.159
|
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr20_-_47238082
|
0.158
|
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr19_-_55914837
|
0.156
|
|
|
|
|
chr13_-_74953873
|
0.155
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr6_+_31698447
|
0.152
|
|
|
|
|
chr1_-_27802688
|
0.150
|
NM_001029882
|
AHDC1
|
AT hook, DNA binding motif, containing 1
|